A Hinge In A Pump Linked To Breast Cancer Survival
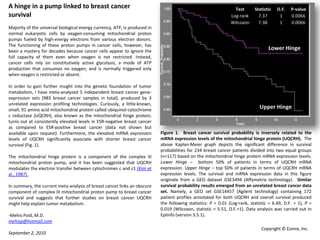
- 1. A hinge in a pump linked to breast cancer survival Majority of the universal biological energy currency, ATP, is produced in normal eukaryotic cells by oxygen-consuming mitochondrial proton pumps fueled by high-energy electrons from various electron donors. The functioning of these proton pumps in cancer cells, however, has been a mystery for decades because cancer cells appear to ignore the full capacity of them even when oxygen is not restricted. Instead, cancer cells rely on constitutively active glycolysis, a mode of ATP production that consumes no oxygen, and is normally triggered only when oxygen is restricted or absent. In order to gain further insight into the genetic foundation of tumor metabolism, I have meta-analyzed 5 independent breast cancer gene-expression sets (983 breast cancer samples in total), produced by 3 unrelated expression profiling technologies. Curiously, a little-known, small, 91 amino acid mitochondrial protein called ubiquinol-cytochrome c reductase (UQCRH), also known as the mitochondrial hinge protein, turns out at consistently elevated levels in ESR-negative breast cancer as compared to ESR-positive breast cancer (data not shown but available upon request). Furthermore, the elevated mRNA expression levels of UQCRH significantly associate with shorter breast cancer survival (Fig. 1). The mitochondrial hinge protein is a component of the complex III mitochondrial proton pump, and it has been suggested that UQCRHmodulates the electron transfer between cytochromes c and c1 (Kim et al., 1987). In summary, the current meta-analysis of breast cancer links an obscure component of complex III mitochondrial proton pump to breast cancer survival and suggests that further studies on breast cancer UQCRH might help explain tumor metabolism. -MehisPold, M.D. mehisp@hotmail.com September 2, 2010 Lower Hinge Upper Hinge Figure 1.Breast cancer survival probability is inversely related to the mRNA expression levels of the mitochondrial hinge protein (UQCRH). The above Kaplan-Meier graph depicts the significant difference in survival probabilities for 234 breast cancer patients divided into two equal groups (n=117) based on the mitochondrial hinge protein mRNA expression levels. Lower Hinge – bottom 50% of patients in terms of UQCRH mRNA expression. Upper Hinge – top 50% of patients in terms of UQCRH mRNA expression levels. The survival and mRNA expression data in this figure originate from a GEO dataset GSE3494 (Affymetrix technology). Similar survival probability results emerged from an unrelated breast cancer data set. Namely, a GEO set GSE14457 (Agilent technology) containing 172 patient profiles annotated for both UQCRH and overall survival produced the following statistics: P = 0.01 (Log-rank, statistic = 6.49, D.F. = 1), P = 0.019 (Wilcoxon, statistic = 5.51, D.F.=1). Data analysis was carried out in EpiInfo (version 3.5.1). Copyright © Eomix, Inc.
