American Statistical Association October 23 2009 Presentation Part 1
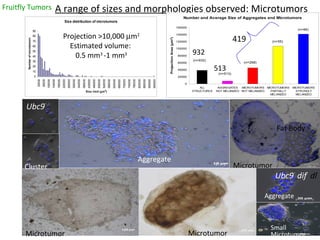
- 1. Fruitfly Tumors A range of sizes and morphologies observed: Microtumors Ubc9 - dif - dl - Ubc9 - - Microtumor Microtumor Microtumor Aggregate Cluster Aggregate Small Microtumor Fat Body 419 Projection >10,000 m 2 Estimated volume: 0.5 mm 3 -1 mm 3 932 513
- 2. (Chiu et al 2005) : dUbc9 negatively regulates the Toll-NF-nB pathways in larval hematopoiesis and drosomycin activation in Drosophila. Developmental Biology. Genotype Number of Larvae Ubc9-(transheterozygote) 58 Bc + Ubc9- 55 95% CI Odds Ratio: NS>5% 0.85- 1.25 Ubc9- Aggregates + Tumors Aggr Tumors Totals 932 513 419 % 55.04% 44.96% Bc Ubc9/+ Ubc9- Aggregates + Tumors Aggr Tumors Totals 874 262 612 % 29.98% 70.02%
- 3. Bc allele background FlyBase GBrowse modENCODE GBrowse Gene Dmelc FB2009_07, released August 10, 2009 General Information Symbol Dmelc Species D. melanogaster Name Black cells Annotation symbol CG5779 Feature type protein_coding_gene FlyBase ID FBgn0000165 Gene Model Status Current Stock availability 68 publicly available Genomic Location Chromosome (arm) 2R Recombination map 2-80.6 Cytogenetic map 54F6-54F6 Sequence location 2R:13,774,718..13,777,477 [-] Genomic Maps The gene Black cells is referred to in FlyBase by the symbol Dmelc (CG5779, FBgn0000165). It is a protein_coding_gene from Drosophila melanogaster. Its sequence location is 2R:13774718..13777477 . It has the cytological map location 54F6 . Its molecular function is described as: monophenol monooxygenase activity; oxygen transporter activity; oxidoreductase activity. It is involved in the biological processes: defense response; melanization defense response; scab formation; response to symbiont; response to wounding; transport. 10 alleles are reported . The phenotypes of these alleles are annotated with: crystal cell; hemocyte; hemolymph; lymph gland; adult; procrystal cell; lamellocyte; posterior lymph gland pair. It has one annotated transcript and one annotated polypeptide . Takehana, A., Katsuyama, T., Yano, T., Oshima, Y., Takada, H., Aigaki, T., Kurata, S. (2002). Overexpression of a pattern-recognition receptor, peptidoglycan-recognition protein-LE, activates imd/relish-mediated antibacterial defense and the prophenoloxidase cascade in Drosophila larvae. Proc. Natl. Acad. Sci. U.S.A. 99(21): 13705--13710. Ye, Y.H., Chenoweth, S.F., McGraw, E.A. (2009). Effective but costly, evolved mechanisms of defense against a virulent opportunistic pathogen in Drosophila melanogaster. PLoS Pathog. 5(4): e1000385.
- 4. Comparative Analysis of Area limits 25K to 300K and 300K to 600K in both Genotypes : Higher Maximum Likelihood mean, variances and wider confidence interval of 25K-300K shows faster mitosis and cell death than that of 300K-600K. Maximum Likelihood (ML) Estimates of BC-All (BC-lwr) and lwr43-5 All BC-All Mean Tumors Variance Tumors 95% Confidence Interval 25K-300K 4.86 0.85 1.22 to 1.84 300K-600K 1.67 0.02 1.11 to 1.20 lwr43-5 All Mean Tumors Variance Tumors 95% Confidence Interval 25K-300K 4.5 0.97 1.10 to 1.88 300K-600K 1.27 0.02 1.05 to 1.12
- 5. 25K-300K Area Size Tumor Log-Normal Distribution in BC-All and Recessive Genotypes (number of micro tumor found or frequency on Y-axis; every 25K scale)
- 6. PROBLEM STATEMENT Tumor size data from non-random and correlated data. Samples were prepared for 8 days and scored on 9 th day- cumulative effects on frequencies of BC-All and recessive (lwr-) Area size Units between 25k to 600k size distributions? Effects of new VS experienced PhD student on data collection? 612 VS 419. This difference is not statistically significant (P> 5%). EXPECTED frequency higher at all area size for Semidominant gene in the hypothetical Y-axis. Does not have a pattern to quantify by a Dynamical simulation equations- tried 100’s of published math methods…. Sample size is ONLY 48 rows of Tumor Frequency data!
- 8. Analysis of Raw data showing V-shape residual and compensatory response by 25K area limit (R-square = 0.36 VS 0.76 VS 0.86 ). Data Analysis needs Dynamical Simulations, Reverse Engineering Algorithms and Simulated OLS Regression.
- 9. LITERATURE REVIEW & METHODS Dynamical Simulation by Taylor’s Power Series like Math equation: A . Y= x 1 + x 2 +x 3 + x 4 . Reference : “Lee Specter and Shawn Luke- Culture Enhances the Evolvability of Cognition. 1996. In Proceedings of the Eighteenth Annual Conference of the Cognitive Science Society. “ According to Specter and Luke, special type of Dynamical Simulation is Symbolic Regression- “ to produce a function, in symbolic form, that fits a provided set of data points. For each element of a set of (x,y) points, the function should map the x value to an appropriate y value. This sort of problem faced by a scientist who has obtained a set of experimental data points and suspects that a simple formula will suffice to explain the data ” . This method is a standard example from Dynamical simulation and used in many different types of biological systems (Koza, J.R. 1992. Genetic Programming: on the programming of computers by means of natural selection. Cambridge, MA, MIT Press).
- 10. B. Reverse Engineering Prediction by the equation of y = 4.251a2 + ln(a2) + 7.243ea- CF . ( Candida Ferreira. 2003. www.gene-expression-programming.com/author.asp- equation 3.2 ) Ekaterina Vladislavleva- June 2008- PhD Theses Models to exhibit not only required properties, but also additional convenient properties like compactness, small number of constants, etc. It is important, that generated models are interpretable and transparent, in order to provide additional understanding of the underlying system or process.
- 12. Reverse Engineering of Polynomial Models of Gene Regulatory Networks (Visual Analytics = Meta Modeling = what are the ranges of input variables that cause the response to take certain values, not necessarily optimal? ) Dr. Eduardo Mendoza Mathematics Department Center for NanoScience Ludwig-Maximilians-University Munich, Germany [email_address] eduardom@math.upd.edu.ph Brody et al . October 1, 2002: PNAS : Significance and Statistical Errors in the analysis of DNA microarray data. 99 (20): 12975-12978 ( Even for Lorentizian like distributions, median of ratios provide distributions more Gaussian like ).
- 13. Reverse Engineering of Systems Systems identification in Engineering: goal is to construct a system with prescribed dynamical properties In Systems Biology, one is interested in identifying as closely as possible a unique biological system that has been observed experimentally In both cases: sparsity of available measurements will leave the system underdetermined (GIGO- Uninterpretable)
- 16. A . “Operon or Tumor Gene Expression occurs in a deterministic way from 25K to 300K area limits, and hence would have high survival probability”. This hypothesis indicates that there are conserved Protein motifs which generates various Brain Tumor sizes in Fruit fly in predetermined frequencies. Thus, micro-tumors counted (frequency) for lower size limits can be predicted by least non-linear mathematical and statistical equations . B . “Log-Normal distribution arose due to compensatory response by lowest size distribution over the next few micro-tumor classes”. If the number of micro-tumors counted for 25K area size is at the expense of next few, then a Log-Normal Distribution can be assured . Log-Normal Distribution explanation
- 17. Leo Breiman: Statist. Sci. Volume 16, Issue 3 (2001), 199-231. Statistical Modeling: The Two Cultures Abstract There are two cultures in the use of statistical modeling to reach conclusions from data. One assumes that the data are generated by a given stochastic data model. The other uses algorithmic models and treats the data mechanism as unknown. The statistical community has been committed to the almost exclusive use of data models. This commitment has led to irrelevant theory, questionable conclusions, and has kept statisticians from working on a large range of interesting current problems. Algorithmic modeling, both in theory and practice, has developed rapidly in fields outside statistics. It can be used both on large complex data sets and as a more accurate and informative alternative to data modeling on smaller data sets. If our goal as a field is to use data to solve problems, then we need to move away from exclusive dependence on data models and adopt a more diverse set of tools .
- 18. A. Analysis of size distribution of lwr (-) microtumors from 58 animals Projection >10,000 m 2 ; Estimated volume: 0.5 mm 3 -1 mm 3 Taylor series: y = x 1 + x 2 +x 3 + x 4 Area Limit Simulated Frequency 100,000 -01 (1) 200,000 +01 (2) 275,000 -02 (3) MLE:25k-300k Mean=4.5 Tumors Variance=0.97 Tumors CI= 1.10-1.88 Tumors MLE: 300k-600k Mean= 1.27 Tumors Variance= 0.02 Tumors CI= 1.05-1.12 Tumors
- 20. Ekaterina Vladislavleva- PhD: JUNE 2008 Both measured and simulated data are very often corrupted by noise, and in case of real measurements can be driven by a combination of both measured and unmeasured input variables, empirical models should not only accurately predict the observed response, but also have some extra generalization capabilities. The same requirement holds for models developed on simulated data. Models to exhibit not only required properties, but also additional convenient properties like compactness, small number of constants, etc. It is important, that generated models are interpretable and transparent, in order to provide additional understanding of the underlying system or process.
