2016 ashg giab poster
•
0 likes•325 views
The document summarizes the Genome in a Bottle Consortium's unprecedented characterization of human reference genomes, including two trios. It describes how the consortium has generated extensive genomic data from various technologies for these genomes and is developing high-confidence calls for variants including SNPs, indels, and structural variants. The consortium is working to characterize more difficult variants and regions, and welcomes new collaborations to further characterize the genomes.
Report
Share
Report
Share
Download to read offline
Recommended
More Related Content
What's hot
What's hot (20)
GIAB Sep2016 Lightning megan cleveland targeted seq

GIAB Sep2016 Lightning megan cleveland targeted seq
The Transforming Genetic Medicine Initiative (TGMI)

The Transforming Genetic Medicine Initiative (TGMI)
171114 best practices for benchmarking variant calls justin

171114 best practices for benchmarking variant calls justin
Aug2015 Ali Bashir and Jason Chin Pac bio giab_assembly_summary_ali3

Aug2015 Ali Bashir and Jason Chin Pac bio giab_assembly_summary_ali3
Genome in a Bottle- reference materials to benchmark challenging variants and...

Genome in a Bottle- reference materials to benchmark challenging variants and...
Similar to 2016 ashg giab poster
Presented by Jennifer D. Churchill, PhD during a special Lunch and Learn session during the American Academy of Forensic Sciences (AAFS) 67th annual conference, February 2015. / Conclusions
• Robust panels of identity and ancestry SNPs
• Robust STR panel
• Whole genome mtDNA sequencing
• Highly informative
• Sensitive
• Quantitative – scaling comparison
• Low density chip is not necessarily a bad chip
• Wide range of density can still yield high quality data
• Based on results continue development and validation
Moving Towards a Validated High Throughput Sequencing Solution for Human Iden...

Moving Towards a Validated High Throughput Sequencing Solution for Human Iden...Thermo Fisher Scientific
Similar to 2016 ashg giab poster (20)
140127 measurements for rm characterization wg summary

140127 measurements for rm characterization wg summary
Randomized Algorithms in Linear Algebra & the Column Subset Selection Problem

Randomized Algorithms in Linear Algebra & the Column Subset Selection Problem
Jan2015 using the pilot genome rm for clinical validation steve lincoln

Jan2015 using the pilot genome rm for clinical validation steve lincoln
RNA-seq: A High-resolution View of the Transcriptome

RNA-seq: A High-resolution View of the Transcriptome
Karen miga centromere sequence characterization and variant detection

Karen miga centromere sequence characterization and variant detection
Moving Towards a Validated High Throughput Sequencing Solution for Human Iden...

Moving Towards a Validated High Throughput Sequencing Solution for Human Iden...
More from GenomeInABottle
More from GenomeInABottle (20)
Using accurate long reads to improve Genome in a Bottle Benchmarks 220923

Using accurate long reads to improve Genome in a Bottle Benchmarks 220923
GIAB Technical Germline Benchmark roadmap discussion

GIAB Technical Germline Benchmark roadmap discussion
GIAB Benchmarks for SVs and Repeats for stanford genetics sv 200511

GIAB Benchmarks for SVs and Repeats for stanford genetics sv 200511
Ga4gh 2019 - Assuring data quality with benchmarking tools from GIAB and GA4GH

Ga4gh 2019 - Assuring data quality with benchmarking tools from GIAB and GA4GH
GIAB GRC Workshop ASHG 2019 Billy Rowell Evaluation of v4 with CCS GATK

GIAB GRC Workshop ASHG 2019 Billy Rowell Evaluation of v4 with CCS GATK
GRC GIAB Workshop ASHG 2019 Small Variant Benchmark

GRC GIAB Workshop ASHG 2019 Small Variant Benchmark
Genome in a Bottle - Towards new benchmarks for the “dark matter” of the huma...

Genome in a Bottle - Towards new benchmarks for the “dark matter” of the huma...
How giab fits in the rest of the world seqc2 tumor normal

How giab fits in the rest of the world seqc2 tumor normal
Recently uploaded
9630942363 THE GENUINE ESCORT AGENCY VIP LUXURY CALL GIRLS
HIGH CLASS MODELS CALL GIRLS GENUINE ESCORT BOOK
BOOK APPOINTMENT - 9630942363 THE GENUINE ESCORT AGENCY
BEST VIP CALL GIRLS & ESCORTS SERVICE 9630942363 VIP CALL GIRLS ALL TYPE WOMEN AVAILABLE
INCALL & OUTCALL BOTH AVAILABLE BOOK NOW
9630942363 VIP GENUINE INDEPENDENT ESCORT AGENCY
VIP PRIVATE AUNTIES
BEAUTIFUL LOOKING HOT AND SEXT GIRLS AND PARTY TYPE GIRLS YOU WANT SERVICE THEN CALL THIS NUMBER 9630942363
ROOM ALSO PROVIDE HOME & HOTELS SERVICE
FULL SAFE AND SECURE WORK
WITHOUT CONDOMS, ORAL, SUCKING, LIP TO LIP, ANAL, BACK SHOTS, SEX 69, WITHOUT BLOWJOB AND MUCH MORE
FOR BOOKING
9630942363Call Girls Ahmedabad Just Call 9630942363 Top Class Call Girl Service Available

Call Girls Ahmedabad Just Call 9630942363 Top Class Call Girl Service AvailableGENUINE ESCORT AGENCY
Model Call Girl Services in Delhi reach out to us at 🔝 9953056974 🔝✔️✔️
Our agency presents a selection of young, charming call girls available for bookings at Oyo Hotels. Experience high-class escort services at pocket-friendly rates, with our female escorts exuding both beauty and a delightful personality, ready to meet your desires. Whether it's Housewives, College girls, Russian girls, Muslim girls, or any other preference, we offer a diverse range of options to cater to your tastes.
We provide both in-call and out-call services for your convenience. Our in-call location in Delhi ensures cleanliness, hygiene, and 100% safety, while our out-call services offer doorstep delivery for added ease.
We value your time and money, hence we kindly request pic collectors, time-passers, and bargain hunters to refrain from contacting us.
Our services feature various packages at competitive rates:
One shot: ₹2000/in-call, ₹5000/out-call
Two shots with one girl: ₹3500/in-call, ₹6000/out-call
Body to body massage with sex: ₹3000/in-call
Full night for one person: ₹7000/in-call, ₹10000/out-call
Full night for more than 1 person: Contact us at 🔝 9953056974 🔝. for details
Operating 24/7, we serve various locations in Delhi, including Green Park, Lajpat Nagar, Saket, and Hauz Khas near metro stations.
For premium call girl services in Delhi 🔝 9953056974 🔝. Thank you for considering us!Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X7![Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X7](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
![Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X7](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X79953056974 Low Rate Call Girls In Saket, Delhi NCR
Recently uploaded (20)
Top Rated Call Girls Kerala ☎ 8250092165👄 Delivery in 20 Mins Near Me

Top Rated Call Girls Kerala ☎ 8250092165👄 Delivery in 20 Mins Near Me
Call Girls Ahmedabad Just Call 9630942363 Top Class Call Girl Service Available

Call Girls Ahmedabad Just Call 9630942363 Top Class Call Girl Service Available
Call Girls Madurai Just Call 9630942363 Top Class Call Girl Service Available

Call Girls Madurai Just Call 9630942363 Top Class Call Girl Service Available
Call Girls Service Jaipur {8445551418} ❤️VVIP BHAWNA Call Girl in Jaipur Raja...

Call Girls Service Jaipur {8445551418} ❤️VVIP BHAWNA Call Girl in Jaipur Raja...
Call Girls Mysore Just Call 8250077686 Top Class Call Girl Service Available

Call Girls Mysore Just Call 8250077686 Top Class Call Girl Service Available
8980367676 Call Girls In Ahmedabad Escort Service Available 24×7 In Ahmedabad

8980367676 Call Girls In Ahmedabad Escort Service Available 24×7 In Ahmedabad
Call Girls Mumbai Just Call 8250077686 Top Class Call Girl Service Available

Call Girls Mumbai Just Call 8250077686 Top Class Call Girl Service Available
Call Girls in Lucknow Just Call 👉👉7877925207 Top Class Call Girl Service Avai...

Call Girls in Lucknow Just Call 👉👉7877925207 Top Class Call Girl Service Avai...
Top Rated Pune Call Girls (DIPAL) ⟟ 8250077686 ⟟ Call Me For Genuine Sex Serv...

Top Rated Pune Call Girls (DIPAL) ⟟ 8250077686 ⟟ Call Me For Genuine Sex Serv...
Call Girls in Delhi Triveni Complex Escort Service(🔝))/WhatsApp 97111⇛47426

Call Girls in Delhi Triveni Complex Escort Service(🔝))/WhatsApp 97111⇛47426
9630942363 Genuine Call Girls In Ahmedabad Gujarat Call Girls Service

9630942363 Genuine Call Girls In Ahmedabad Gujarat Call Girls Service
Low Rate Call Girls Bangalore {7304373326} ❤️VVIP NISHA Call Girls in Bangalo...

Low Rate Call Girls Bangalore {7304373326} ❤️VVIP NISHA Call Girls in Bangalo...
Call Girls Jaipur Just Call 9521753030 Top Class Call Girl Service Available

Call Girls Jaipur Just Call 9521753030 Top Class Call Girl Service Available
Independent Call Girls In Jaipur { 8445551418 } ✔ ANIKA MEHTA ✔ Get High Prof...

Independent Call Girls In Jaipur { 8445551418 } ✔ ANIKA MEHTA ✔ Get High Prof...
Call Girls Rishikesh Just Call 8250077686 Top Class Call Girl Service Available

Call Girls Rishikesh Just Call 8250077686 Top Class Call Girl Service Available
Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X7![Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X7](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
![Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X7](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X7
Coimbatore Call Girls in Coimbatore 7427069034 genuine Escort Service Girl 10...

Coimbatore Call Girls in Coimbatore 7427069034 genuine Escort Service Girl 10...
Call Girls Kolkata Kalikapur 💯Call Us 🔝 8005736733 🔝 💃 Top Class Call Girl Se...

Call Girls Kolkata Kalikapur 💯Call Us 🔝 8005736733 🔝 💃 Top Class Call Girl Se...
💕SONAM KUMAR💕Premium Call Girls Jaipur ↘️9257276172 ↙️One Night Stand With Lo...

💕SONAM KUMAR💕Premium Call Girls Jaipur ↘️9257276172 ↙️One Night Stand With Lo...
2016 ashg giab poster
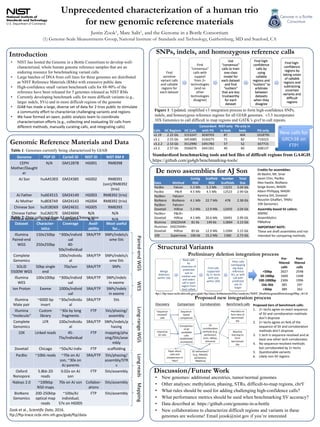
- 1. Introduction Unprecedented characterization of a human trio for new genomic reference materials Justin Zook1, Marc Salit1, and the Genome in a Bottle Consortium (1) Genome-Scale Measurements Group, National Institute of Standards and Technology, Gaithersburg, MD and Stanford, CA • NIST has hosted the Genome in a Bottle Consortium to develop well- characterized, whole human genome reference samples that are an enduring resource for benchmarking variant calls • Large batches of DNA from cell lines for these genomes are distributed as NIST Reference Materials (RMs) with extensive public data • High-confidence small variant benchmark calls for 88-90% of the reference have been released for 5 genomes released as NIST RMs • Currently developing benchmark calls for more difficult variants (e.g., larger indels, SVs) and in more difficult regions of the genome • GIAB has made a large, diverse set of data for 2 trios public to s:mulate a community effort to characterize challenging variants and regions • We have formed an open, public analysis team to coordinate characteriza:on efforts (e.g., collec:ng and evalua:ng SV calls from different methods, manually cura:ng calls, and integra:ng calls) Genomic Reference Materials and Data Figure 1: Updated, simplified v3 integration process to form high-confidence SNPs, indels, and homozygous reference regions for all GIAB genomes. v3.3 incorporates 10X Genomics to call difficult to map regions and GATK’s gvcf to call repeats. Discussion/Future Work • New genomes: additional ancestries, tumor/normal genomes • Other analyses: methylation, phasing, STRs, difficult-to-map regions, chrY • What rules should be used for adding challenging high-confidence calls? • What performance metrics should be used when benchmarking SV accuracy? • Data described at: https://github.com/genome-in-a-bottle • New collaborations to characterize difficult regions and variants in these genomes are welcome! Email jzook@nist.gov if you’re interested Genome PGP ID Coriell ID NIST ID NIST RM # CEPH Mother/ Daughter N/A GM12878 HG001 RM8398 AJ Son huAA53E0 GM24385 HG002 RM8391 (son)/ RM8392 (trio) AJ Father hu6E4515 GM24149 HG003 RM8392 (trio) AJ Mother hu8E87A9 GM24143 HG004 RM8392 (trio) Chinese Son hu91BD69 GM24631 HG005 RM8393 Chinese Father huCA017E GM24694 N/A N/A Chinese Mother hu38168C GM24695 N/A N/A Dataset Character- is:cs Coverage Avail- ability Most useful for… Illumina Paired-end WGS 150x150bp 250x250bp ~300x/ individual 40-50x/ individual SRA/FTP SNPs/indels/ some SVs Complete Genomics 100x/ individual SRA/FTP SNPs/indels/ some SVs SOLiD 5500W WGS 50bp single end 70x/son SRA/FTP SNPs Illumina WES 100x100bp ~300x/ individual SRA/FTP SNPs/indels in exome Ion Proton Exome 1000x/ individual SRA/FTP SNPs/indels in exome Illumina Mate pair ~6000 bp insert ~30x/ individual SRA/FTP SVs Illumina “moleculo” Custom library ~30x by long fragments FTP SVs/phasing/ assembly Complete Genomics LFR 100x/ individual SRA/FTP SNPs/indels/ phasing 10X Linked reads 45-75x/ individual FTP mapping/ phasing/SVs/ assembly Dovetail Chicago ~50x/AJ indiv FTP scaffolding PacBio ~10kb reads ~70x on AJ son, ~30x on AJ parents SRA/FTP SVs/phasing/ assembly/ STRs Oxford Nanopore 5.8kb 2D reads 0.02x on AJ son FTP SVs/assembly Nabsys 2.0 ~100kbp N50 maps 70x on AJ son Collabor- a:ons SVs/assembly BioNano Genomics 200-250kbp op:cal map reads ~100x/AJ individual; 57x on HG005 FTP SVs/assembly Long-range WGS WES Long reads Mapping Paired-end WGS De novo assemblies for AJ Son SNPs, indels, and homozygous reference calls Data Method Con:g N50 Scaffold N50 Number Scaffolds Total Size PacBio Falcon 5.3 Mb 5.3 Mb 13231 3.04 Gb PacBio PBcR 4.5 Mb 4.5 Mb 12523 2.99 Gb PacBio+ BioNano Falcon+ BioNano 4.1 Mb 22.7 Mb 478 2.38 Gb PacBio+ Dovetail Falcon+ HiRise 5.3 Mb 12.9 Mb 12459 3.04 Gb PacBio+ Dovetail PBcR+ HiRise 4.1 Mb 20.6 Mb 10491 2.99 Gb Illumina DISCOVAR 81 kb 149 kb 1.06M 3.13 Gb Illumina+ Dovetail DISCOVAR +HiRise 85 kb 12.9 Mb 1.03M 3.15 Gb 10X Supernova 106 kb 15.2 Mb 1360 2.73 Gb Find sensi:ve variant calls and callable regions for each dataset Find “consensus” calls with support from 2+ technologies (and no other technologies disagree) Use “consensus” calls to train one-class model for each dataset and find “outliers” that are less trustworthy for each dataset Find high- confidence calls by using callable regions and “outliers” to arbitrate between datasets when they disagree Find high- confidence regions by taking union of callable regions and subtrac:ng uncertain variants and difficult regions Table 1: Genomes currently being characterized by GIAB Table 2: Data collected from AJ and/or Chinese trios Credits for assemblies: Ali Bashir, Mt. Sinai Jason Chin, PacBio Alex Has:e, BioNano Serge Koren, NHGRI Adam Phillippy, NHGRI Kareina Dill, Dovetail Noushin Ghaffari, TAMU 10X Genomics Zook et al., Scien&fic Data, 2016. kp://kp-trace.ncbi.nlm.nih.gov/giab/kp/data “sequence- resolved” calls Discovery Imprecise SV calls Sequence- based comparison SV corrobora:on methods (e.g., parliament, svviz, nabsys, bionano) Heuris:cs to form :ers of benchmark SVs Machine learning to form benchmark SVs Comparison of all candidate calls (SURVIVOR/ svcompare) Comparison Corrobora:on Benchmark calls SV refinement? (e.g., MetaSV, parliament, PBRefine) Paper about calls and comparisons in ~Nov? Structural Variants Calls HC Regions HC Calls Concordant with PG NIST-only in beds PG-only in beds PG-only v2.19 2.22 Gb 3153247 3030703 87 404 1018795 v3.1 2.55 Gb 3453085 3330275 71 82 719223 v3.2.2 2.53 Gb 3512990 3391783 57 52 657715 v3.3 2.57 Gb 3566076 3441361 40 60 608137 Proposed new integration process Proposed :ers of benchmark calls: 1. 2+ techs agree on exact sequence of SV and corrobora:on methods don’t disprove 2. 2+ techs agree on 9x% of the sequence of SV and corrobora:on methods don’t disprove 3. 1 tech is sequence-resolved and at least one other tech corroborates 4. No sequence-resolved methods but corroborated by 2+ techs 5. Ques:onable variants 6. Likely non-SV regions New calls for GRCh38 on FTP! Merge dele:ons within 1kb Rank calls by closeness of predicted size to median size and select call in each region from best callset Find calls supported by 2+ techs with size within 20% Filter calls overlapping seg dups, reference N’s, or with call with predicted size 2x larger Preliminary deletion integration process Pre- filtered calls Post- filtered calls <50bp 2627 2548 50-100bp 1600 1448 100-1000bp 2306 1996 1kb-3kb 385 297 >3kbp 389 262 ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data/AshkenazimTrio/analysis/NIST_DraftIntegratedDeletionsgt19bp_v0.1.8 Standardized benchmarking tools and bed files of difficult regions from GA4GH: https://github.com/ga4gh/benchmarking-tools/ Assembly-based SV callers: MSPAC Assembly:cs PBRefine IMPORTANT NOTE: These are drak assemblies and not intended for comparing methods.
