Giab agbt SVs_2019
•
3 j'aime•22,429 vues
AGBT poster describing structural variant (SV) benchmark
Signaler
Partager
Signaler
Partager
Télécharger pour lire hors ligne
Recommandé
Recommandé
Alzheimer’s disease (AD) is a devastating neurodegenerative disease that is genetically complex. Although great progress has been made in identifying fully penetrant mutations in genes that cause early-onset AD, these still represent a very small percentage of AD cases. Large-scale, genome-wide association studies (GWAS) have identified at least 20 additional genetic risk loci for the more common form: late-onset AD. However, the identified SNPs are typically not the actual risk variants, but are in linkage disequilibrium with the presumed causative variants [1].
To help identify causative genetic variants, we have combined highly accurate, long-read sequencing with hybrid-capture technology. In this collaborative webinar*, we present this method and show how combining IDT xGen® Lockdown® Probes with PacBio SMRT® Sequencing allows targeting and sequencing of candidate genes from genomic DNA and corresponding transcripts from cDNA. Using a panel of target capture probes for 35 AD candidate genes, we demonstrate the power of this approach by looking at data for two individuals with AD. Some additional benefits of this method include the ability to leverage long reads, phase heterozygous variants, and link corresponding transcript isoforms to their respective alleles.
Reference: 1. Van Cauwenberghe C, Van Broeckhoven C, Sleegers K. (2016) The genetic landscape of Alzheimer disease: clinical implications and perspectives. Genet Med, 18(5):421–430.
* This presentation represents a collaboration between Pacific Biosciences and Integrated DNA Technologies. The individual opinions expressed may not reflect shared opinions of Pacific Biosciences and Integrated DNA Technologies.Characterizing Alzheimer’s Disease candidate genes and transcripts with targe...

Characterizing Alzheimer’s Disease candidate genes and transcripts with targe...Integrated DNA Technologies
CRISPR has become an increasingly popular tool for genome editing, in part because it is highly flexible and relatively easy to implement compared to other technologies. However, for scientists beginning to work with this method, the wide range of products and variety of editing approaches can be overwhelming. In this presentation, Justin Barr provides a simple explanation of the steps for planning your experiment, including guide RNA design, an overview of delivery methods, and options for measuring editing results. He also discusses how to generate specific mutations in the genome using homology-directed repair (HDR).Getting started with CRISPR: a review of gene knockout and homology-directed ...

Getting started with CRISPR: a review of gene knockout and homology-directed ...Integrated DNA Technologies
Contenu connexe
Tendances
Alzheimer’s disease (AD) is a devastating neurodegenerative disease that is genetically complex. Although great progress has been made in identifying fully penetrant mutations in genes that cause early-onset AD, these still represent a very small percentage of AD cases. Large-scale, genome-wide association studies (GWAS) have identified at least 20 additional genetic risk loci for the more common form: late-onset AD. However, the identified SNPs are typically not the actual risk variants, but are in linkage disequilibrium with the presumed causative variants [1].
To help identify causative genetic variants, we have combined highly accurate, long-read sequencing with hybrid-capture technology. In this collaborative webinar*, we present this method and show how combining IDT xGen® Lockdown® Probes with PacBio SMRT® Sequencing allows targeting and sequencing of candidate genes from genomic DNA and corresponding transcripts from cDNA. Using a panel of target capture probes for 35 AD candidate genes, we demonstrate the power of this approach by looking at data for two individuals with AD. Some additional benefits of this method include the ability to leverage long reads, phase heterozygous variants, and link corresponding transcript isoforms to their respective alleles.
Reference: 1. Van Cauwenberghe C, Van Broeckhoven C, Sleegers K. (2016) The genetic landscape of Alzheimer disease: clinical implications and perspectives. Genet Med, 18(5):421–430.
* This presentation represents a collaboration between Pacific Biosciences and Integrated DNA Technologies. The individual opinions expressed may not reflect shared opinions of Pacific Biosciences and Integrated DNA Technologies.Characterizing Alzheimer’s Disease candidate genes and transcripts with targe...

Characterizing Alzheimer’s Disease candidate genes and transcripts with targe...Integrated DNA Technologies
CRISPR has become an increasingly popular tool for genome editing, in part because it is highly flexible and relatively easy to implement compared to other technologies. However, for scientists beginning to work with this method, the wide range of products and variety of editing approaches can be overwhelming. In this presentation, Justin Barr provides a simple explanation of the steps for planning your experiment, including guide RNA design, an overview of delivery methods, and options for measuring editing results. He also discusses how to generate specific mutations in the genome using homology-directed repair (HDR).Getting started with CRISPR: a review of gene knockout and homology-directed ...

Getting started with CRISPR: a review of gene knockout and homology-directed ...Integrated DNA Technologies
Tendances (20)
How giab fits in the rest of the world seqc2 tumor normal

How giab fits in the rest of the world seqc2 tumor normal
Characterizing Alzheimer’s Disease candidate genes and transcripts with targe...

Characterizing Alzheimer’s Disease candidate genes and transcripts with targe...
New RNA tools for optimized CRISPR/Cas9 genome editing

New RNA tools for optimized CRISPR/Cas9 genome editing
Getting started with CRISPR: a review of gene knockout and homology-directed ...

Getting started with CRISPR: a review of gene knockout and homology-directed ...
Genome in a bottle for ashg grc giab workshop 181016

Genome in a bottle for ashg grc giab workshop 181016
Similaire à Giab agbt SVs_2019
Course: Bioinformatics for Biomedical Research (2014).
Session: 2.1.3- Next Generation Sequencing. Technologies and Applications. Part III: NGS Applications II.
Statistics and Bioinformatisc Unit (UEB) & High Technology Unit (UAT) from Vall d'Hebron Research Institute (www.vhir.org), Barcelona.NGS Applications II (UEB-UAT Bioinformatics Course - Session 2.1.3 - VHIR, Ba...

NGS Applications II (UEB-UAT Bioinformatics Course - Session 2.1.3 - VHIR, Ba...VHIR Vall d’Hebron Institut de Recerca
Similaire à Giab agbt SVs_2019 (20)
GIAB Benchmarks for SVs and Repeats for stanford genetics sv 200511

GIAB Benchmarks for SVs and Repeats for stanford genetics sv 200511
2014 June 17 PacBio User Group Meeting Presentation "How Looking for a Needle...

2014 June 17 PacBio User Group Meeting Presentation "How Looking for a Needle...
NGS Applications II (UEB-UAT Bioinformatics Course - Session 2.1.3 - VHIR, Ba...

NGS Applications II (UEB-UAT Bioinformatics Course - Session 2.1.3 - VHIR, Ba...
Towards Precision Medicine: Tute Genomics, a cloud-based application for anal...

Towards Precision Medicine: Tute Genomics, a cloud-based application for anal...
Microbiome studies using 16S ribosomal DNA PCR: some cautionary tales.

Microbiome studies using 16S ribosomal DNA PCR: some cautionary tales.
Expanding Your Research Capabilities Using Targeted NGS

Expanding Your Research Capabilities Using Targeted NGS
Plus de GenomeInABottle
Plus de GenomeInABottle (15)
Using accurate long reads to improve Genome in a Bottle Benchmarks 220923

Using accurate long reads to improve Genome in a Bottle Benchmarks 220923
Genome in a Bottle- reference materials to benchmark challenging variants and...

Genome in a Bottle- reference materials to benchmark challenging variants and...
GIAB Technical Germline Benchmark roadmap discussion

GIAB Technical Germline Benchmark roadmap discussion
GIAB GRC Workshop ASHG 2019 Billy Rowell Evaluation of v4 with CCS GATK

GIAB GRC Workshop ASHG 2019 Billy Rowell Evaluation of v4 with CCS GATK
GRC GIAB Workshop ASHG 2019 Small Variant Benchmark

GRC GIAB Workshop ASHG 2019 Small Variant Benchmark
New data from giab genomes intro and ultralong nanopore

New data from giab genomes intro and ultralong nanopore
How giab fits in the rest of the world mdic somatic reference samples

How giab fits in the rest of the world mdic somatic reference samples
How giab fits in the rest of the world telomere to telomere consortium

How giab fits in the rest of the world telomere to telomere consortium
How giab fits in the rest of the world human genome structural variation co...

How giab fits in the rest of the world human genome structural variation co...
Dernier
Model Call Girl Services in Delhi reach out to us at 🔝 9953056974 🔝✔️✔️
Our agency presents a selection of young, charming call girls available for bookings at Oyo Hotels. Experience high-class escort services at pocket-friendly rates, with our female escorts exuding both beauty and a delightful personality, ready to meet your desires. Whether it's Housewives, College girls, Russian girls, Muslim girls, or any other preference, we offer a diverse range of options to cater to your tastes.
We provide both in-call and out-call services for your convenience. Our in-call location in Delhi ensures cleanliness, hygiene, and 100% safety, while our out-call services offer doorstep delivery for added ease.
We value your time and money, hence we kindly request pic collectors, time-passers, and bargain hunters to refrain from contacting us.
Our services feature various packages at competitive rates:
One shot: ₹2000/in-call, ₹5000/out-call
Two shots with one girl: ₹3500/in-call, ₹6000/out-call
Body to body massage with sex: ₹3000/in-call
Full night for one person: ₹7000/in-call, ₹10000/out-call
Full night for more than 1 person: Contact us at 🔝 9953056974 🔝. for details
Operating 24/7, we serve various locations in Delhi, including Green Park, Lajpat Nagar, Saket, and Hauz Khas near metro stations.
For premium call girl services in Delhi 🔝 9953056974 🔝. Thank you for considering us!Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X7![Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X7](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
![Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X7](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X79953056974 Low Rate Call Girls In Saket, Delhi NCR
9630942363 THE GENUINE ESCORT AGENCY VIP LUXURY CALL GIRLS
HIGH CLASS MODELS CALL GIRLS GENUINE ESCORT BOOK
BOOK APPOINTMENT - 9630942363 THE GENUINE ESCORT AGENCY
BEST VIP CALL GIRLS & ESCORTS SERVICE 9630942363 VIP CALL GIRLS ALL TYPE WOMEN AVAILABLE
INCALL & OUTCALL BOTH AVAILABLE BOOK NOW
9630942363 VIP GENUINE INDEPENDENT ESCORT AGENCY
VIP PRIVATE AUNTIES
BEAUTIFUL LOOKING HOT AND SEXT GIRLS AND PARTY TYPE GIRLS YOU WANT SERVICE THEN CALL THIS NUMBER 9630942363
ROOM ALSO PROVIDE HOME & HOTELS SERVICE
FULL SAFE AND SECURE WORK
WITHOUT CONDOMS, ORAL, SUCKING, LIP TO LIP, ANAL, BACK SHOTS, SEX 69, WITHOUT BLOWJOB AND MUCH MORE
FOR BOOKING
9630942363Call Girls Ahmedabad Just Call 9630942363 Top Class Call Girl Service Available

Call Girls Ahmedabad Just Call 9630942363 Top Class Call Girl Service AvailableGENUINE ESCORT AGENCY
🌹Attapur⬅️ Vip Call Girls Hyderabad 📱9352852248 Book Well Trand Call Girls In Hyderabad Escorts Service
Escorts Service Available
Whatsapp Chaya ☎️ : [+91-9352852248 ]
Escorts Service Hyderabad are always ready to make their clients happy. Their exotic looks and sexy personalities are sure to turn heads. You can enjoy with them, including massages and erotic encounters.#P12Our area Escorts are young and sexy, so you can expect to have an exotic time with them. They are trained to satiate your naughty nerves and they can handle anything that you want. They are also intelligent, so they know how to make you feel comfortable and relaxed
SERVICE ✅ ❣️
⭐➡️HOT & SEXY MODELS // COLLEGE GIRLS HOUSE WIFE RUSSIAN , AIR HOSTES ,VIP MODELS .
AVAILABLE FOR COMPLETE ENJOYMENT WITH HIGH PROFILE INDIAN MODEL AVAILABLE HOTEL & HOME
★ SAFE AND SECURE HIGH CLASS SERVICE AFFORDABLE RATE
★
SATISFACTION,UNLIMITED ENJOYMENT.
★ All Meetings are confidential and no information is provided to any one at any cost.
★ EXCLUSIVE PROFILes Are Safe and Consensual with Most Limits Respected
★ Service Available In: - HOME & HOTEL Star Hotel Service .In Call & Out call
SeRvIcEs :
★ A-Level (star escort)
★ Strip-tease
★ BBBJ (Bareback Blowjob)Receive advanced sexual techniques in different mode make their life more pleasurable.
★ Spending time in hotel rooms
★ BJ (Blowjob Without a Condom)
★ Completion (Oral to completion)
★ Covered (Covered blowjob Without condom
★ANAL SERVICES.
🌹Attapur⬅️ Vip Call Girls Hyderabad 📱9352852248 Book Well Trand Call Girls In...

🌹Attapur⬅️ Vip Call Girls Hyderabad 📱9352852248 Book Well Trand Call Girls In...Call Girls In Delhi Whatsup 9873940964 Enjoy Unlimited Pleasure
Dernier (20)
Andheri East ) Call Girls in Mumbai Phone No 9004268417 Elite Escort Service ...

Andheri East ) Call Girls in Mumbai Phone No 9004268417 Elite Escort Service ...
Call Girls Service Jaipur {9521753030 } ❤️VVIP BHAWNA Call Girl in Jaipur Raj...

Call Girls Service Jaipur {9521753030 } ❤️VVIP BHAWNA Call Girl in Jaipur Raj...
Call Girls in Delhi Triveni Complex Escort Service(🔝))/WhatsApp 97111⇛47426

Call Girls in Delhi Triveni Complex Escort Service(🔝))/WhatsApp 97111⇛47426
Call Girls Kolkata Kalikapur 💯Call Us 🔝 8005736733 🔝 💃 Top Class Call Girl Se...

Call Girls Kolkata Kalikapur 💯Call Us 🔝 8005736733 🔝 💃 Top Class Call Girl Se...
Low Rate Call Girls Bangalore {7304373326} ❤️VVIP NISHA Call Girls in Bangalo...

Low Rate Call Girls Bangalore {7304373326} ❤️VVIP NISHA Call Girls in Bangalo...
Coimbatore Call Girls in Thudiyalur : 7427069034 High Profile Model Escorts |...

Coimbatore Call Girls in Thudiyalur : 7427069034 High Profile Model Escorts |...
Call Girls Amritsar Just Call 8250077686 Top Class Call Girl Service Available

Call Girls Amritsar Just Call 8250077686 Top Class Call Girl Service Available
VIP Hyderabad Call Girls Bahadurpally 7877925207 ₹5000 To 25K With AC Room 💚😋

VIP Hyderabad Call Girls Bahadurpally 7877925207 ₹5000 To 25K With AC Room 💚😋
Most Beautiful Call Girl in Bangalore Contact on Whatsapp

Most Beautiful Call Girl in Bangalore Contact on Whatsapp
Call Girls Varanasi Just Call 8250077686 Top Class Call Girl Service Available

Call Girls Varanasi Just Call 8250077686 Top Class Call Girl Service Available
Premium Call Girls In Jaipur {8445551418} ❤️VVIP SEEMA Call Girl in Jaipur Ra...

Premium Call Girls In Jaipur {8445551418} ❤️VVIP SEEMA Call Girl in Jaipur Ra...
Call Girls Service Jaipur {9521753030} ❤️VVIP RIDDHI Call Girl in Jaipur Raja...

Call Girls Service Jaipur {9521753030} ❤️VVIP RIDDHI Call Girl in Jaipur Raja...
Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X7![Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X7](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
![Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X7](data:image/gif;base64,R0lGODlhAQABAIAAAAAAAP///yH5BAEAAAAALAAAAAABAAEAAAIBRAA7)
Call Girls in Gagan Vihar (delhi) call me [🔝 9953056974 🔝] escort service 24X7
Call Girls Ahmedabad Just Call 9630942363 Top Class Call Girl Service Available

Call Girls Ahmedabad Just Call 9630942363 Top Class Call Girl Service Available
Coimbatore Call Girls in Coimbatore 7427069034 genuine Escort Service Girl 10...

Coimbatore Call Girls in Coimbatore 7427069034 genuine Escort Service Girl 10...
Call Girl In Pune 👉 Just CALL ME: 9352988975 💋 Call Out Call Both With High p...

Call Girl In Pune 👉 Just CALL ME: 9352988975 💋 Call Out Call Both With High p...
🌹Attapur⬅️ Vip Call Girls Hyderabad 📱9352852248 Book Well Trand Call Girls In...

🌹Attapur⬅️ Vip Call Girls Hyderabad 📱9352852248 Book Well Trand Call Girls In...
(Low Rate RASHMI ) Rate Of Call Girls Jaipur ❣ 8445551418 ❣ Elite Models & Ce...

(Low Rate RASHMI ) Rate Of Call Girls Jaipur ❣ 8445551418 ❣ Elite Models & Ce...
Top Rated Hyderabad Call Girls Chintal ⟟ 9332606886 ⟟ Call Me For Genuine Se...

Top Rated Hyderabad Call Girls Chintal ⟟ 9332606886 ⟟ Call Me For Genuine Se...
Call Girls Hyderabad Just Call 8250077686 Top Class Call Girl Service Available

Call Girls Hyderabad Just Call 8250077686 Top Class Call Girl Service Available
Giab agbt SVs_2019
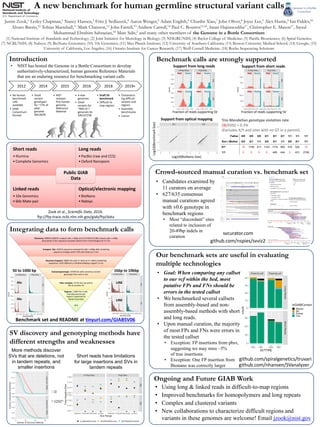
- 1. Discovery: 498876 (296761 unique) calls >=50bp and 1157458 (521360 unique) calls >=20bp discovered in 30+ sequence-resolved callsets from 4 technologies for AJ Trio Compare SVs: 128715 sequence-resolved SV calls >=50bp after clustering sequence changes within 20% edit distance in trio Discovery Support: 30062 SVs with 2+ techs or 5+ callers predicting sequences <20% different or BioNano/Nabsys support in trio Evaluate/genotype: 19748 SVs with consensus variant genotype from svviz in son Filter complex: 12745 SVs not within 1kb of another SV Regions: 11869 SVs inside 2.69 Gbp benchmark regions supported by diploid assembly v0.6 Introduction A new benchmark for human germline structural variant calls Justin Zook,1 Lesley Chapman,1 Nancy Hansen,3 Fritz J. Sedlazeck,4 Aaron Wenger,5 Adam English,6 Chunlin Xiao,7 John Oliver,8 Joyce Lee,9 Alex Hastie,9 Ian Fiddes,10 Alvaro Barrio,10 Tobias Marschall,11 Mark Chaisson,12 John Farrell,13 Andrew Carroll,14 Paul C. Boutros15,16, Iman Hajirasouliha17, Christopher E. Mason17, Sayed Mohammad Ebrahim Sahraeian,18 Marc Salit,2 and many other members of the Genome in a Bottle Consortium (1) National Institute of Standards and Technology; (2) Joint Initiative for Metrology in Biology; (3) NHGRI/NIH; (4) Baylor College of Medicine; (5) Pacific Biosciences; (6) Spiral Genetics; (7) NCBI/NIH; (8) Nabsys; (9) BioNano Genomics; (10) 10x Genomics; (11) Max Planck Institute; (12) University of Southern California; (13) Boston University Medical School; (14) Google; (15) University of California, Los Angeles; (16) Ontario Institute for Cancer Research; (17) Weill Cornell Medicine; (18) Roche Sequencing Solutions • NIST has hosted the Genome in a Bottle Consortium to develop authoritatively-characterized, human genome Reference Materials that are an enduring resource for benchmarking variant calls Integrating data to form benchmark calls Ongoing and Future GIAB Work • Using long & linked reads in difficult-to-map regions • Improved benchmarks for homopolymers and long repeats • Complex and clustered variants • New collaborations to characterize difficult regions and variants in these genomes are welcome! Email jzook@nist.gov Crowd-sourced manual curation vs. benchmark set Benchmark calls are strongly supported Zook et al., Scientific Data, 2016. ftp://ftp-trace.ncbi.nlm.nih.gov/giab/ftp/data Our benchmark sets are useful in evaluating multiple technologies 2012 • No human benchmark calls available • GIAB Consortium formed 2014 • Small variant genotypes for ~77% of pilot genome NA12878 2015 • NIST releases first human genome Reference Material 2016 • 4 new genomes • Small variants for ~90% of 7 genomes for GRCh37/38 2018 • Draft SV benchmark • Difficult to map regions 2019+ • Characteriz- ing difficult variants and regions • Assembly benchmarks • Cancer Benchmark set and README at tinyurl.com/GIABSV06 • Goal: When comparing any callset to our vcf within the bed, most putative FPs and FNs should be errors in the tested callset • We benchmarked several callsets from assembly-based and non- assembly-based methods with short and long reads. • Upon manual curation, the majority of most FPs and FNs were errors in the tested callset • Exception: FP insertions from pbsv, suggesting we may miss ~5% of true insertions • Exception: One FP insertion from Bionano was correctly larger github.com/nspies/svviz2 50 to 1000 bp svcurator.com 1kbp to 10kbp Alu Alu LINE LINE • Candidates examined by 11 curators on average • 627/635 consensus manual curations agreed with v0.6 genotype in benchmark regions • Most “discordant” sites related to inclusion of 20-49bp indels in curation github.com/spiralgenetics/truvari Short reads • Illumina • Complete Genomics Long reads • PacBio (raw and CCS) • Oxford Nanopore Linked reads • 10x Genomics • 6kb Mate-pair Optical/electronic mapping • BioNano • Nabsys Public GIAB Data Short reads have limitations for large insertions and SVs in tandem repeats Log10(BioNano Size) Log10(BenchmarkSize) Father 0/0 0/0 0/0 0/1 0/1 0/1 1/1 1/1 1/1 Son | Mother 0/0 0/1 1/1 0/0 0/1 1/1 0/0 0/1 1/1 0/1 14 1185 417 1143 1119 462 416 522 12 1/1 0 0 0 0 449 444 2 431 2748 Trio Mendelian genotype violation rate 28/9392 = 0.3% (Excludes X/Y and sites with no GT in a parent) Support from long reads Support from short reads Support from optical mapping SV discovery and genotyping methods have different strengths and weaknesses More methods discover SVs that are deletions, not in tandem repeats, and smaller insertions github.com/nhansen/SVanalyzer Fraction of reads supporting SV Fraction of reads supporting SV Het Hom Het Hom Het Hom Het Hom Het Hom Het Hom Het Hom Het Hom
