Eukaryotic transcription
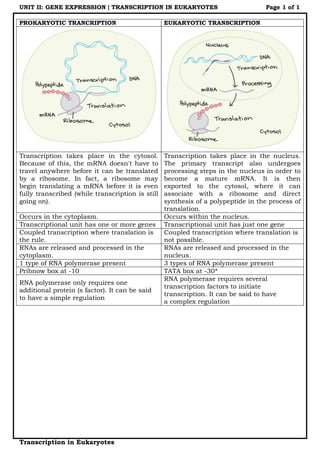
- 1. UNIT II: GENE EXPRESSION | TRANSCRIPTION IN EUKARYOTES Page 1 of 1 Transcription in Eukaryotes PROKARYOTIC TRANCRIPTION EUKARYOTIC TRANSCRIPTION Transcription takes place in the cytosol. Because of this, the mRNA doesn't have to travel anywhere before it can be translated by a ribosome. In fact, a ribosome may begin translating a mRNA before it is even fully transcribed (while transcription is still going on). Transcription takes place in the nucleus. The primary transcript also undergoes processing steps in the nucleus in order to become a mature mRNA. It is then exported to the cytosol, where it can associate with a ribosome and direct synthesis of a polypeptide in the process of translation. Occurs in the cytoplasm. Occurs within the nucleus. Transcriptional unit has one or more genes Transcriptional unit has just one gene Coupled transcription where translation is the rule. Coupled transcription where translation is not possible. RNAs are released and processed in the cytoplasm. RNAs are released and processed in the nucleus. 1 type of RNA polymerase present 3 types of RNA polymerase present Pribnow box at -10 TATA box at -30* RNA polymerase only requires one additional protein (s factor). It can be said to have a simple regulation RNA polymerase requires several transcription factors to initiate transcription. It can be said to have a complex regulation
- 2. UNIT II: GENE EXPRESSION | TRANSCRIPTION IN EUKARYOTES Page 1 of 1 Transcription in Eukaryotes TRANSCRIPTION IN EUKARYOTES Transcription is more complicated in eukaryotes than in prokaryotes because eukaryotes possess three different classes of RNA polymerases and because of the way in which transcripts are processed to their functional forms. More proteins and transcription factors are involved in eukaryotic transcription. EUKARYOTIC RNA POLYMERASES In eukaryotes, three different RNA polymerases transcribe the genes for 4 types of RNA’s. RNA polymerases in eukaryotes consist of multiple subunits. For example: In Yeast, RNA polymerase II consists of 12 subunits and has a U - shaped structure. The larger subunit of polymerase II has a Carboxy-Terminal Domain (CTD) which extends as a tail. The CTD contains a series of repeats of the heptapeptide sequence; Tyr-ser-pro-Thr-ser-pro-ser. These repeats contain sites for phosphorylation by specific kinase. This phosphorylation has a major role in elongation phase. TYPES OF EUKARYOTIC RNA POLYMERASES ENZYME LOCATION FUNCTION 1 RNA polymerase I Nucleolus Synthesis of 3 subunits of the RNAs found in the ribosomes: 28s,18s and 5.8s 2 RNA polymerase II Nucleoplasm Synthesis of messenger RNA (mRNA) and some small nuclear RNA (snRNA) 3 RNA polymerase III Nucleoplasm Synthesis of transfer RNA (tRNA), 5s rRNA & snRNAs which are not made by RNA pol 2 In eukaryotes there are three RNA polymerases: I, II and III. The process includes a proofreading mechanism. RNA polymerase I is located in the nucleolus, a specialized nuclear substructure in which ribosomal RNA (rRNA) is transcribed, processed, and assembled into ribosomes. The rRNA molecules are considered structural RNAs because they have a cellular role but are not translated into protein. The rRNAs are components of the ribosome and are essential to the process of translation. RNA polymerase I synthesizes all of the rRNAs except for the 5S rRNA molecule. RNA polymerase II is located in the nucleus and synthesizes all protein-coding nuclear pre-mRNAs. Eukaryotic pre-mRNAs undergo extensive processing after transcription. RNA polymerase II is responsible for transcribing the overwhelming majority of eukaryotic genes, including all of the protein-encoding genes which ultimately are translated into proteins and genes for several types of regulatory RNAs, including microRNAs (miRNAs) and long-coding RNAs (lncRNAs). RNA polymerase III is also located in the nucleus. This polymerase transcribes a variety of structural RNAs that includes the 5S pre-rRNA, transfer pre-RNAs (pre- tRNAs), and small nuclear pre-RNAs. The tRNAs have a critical role in translation: they serve as the adaptor molecules between the mRNA template and the growing polypeptide chain. Small nuclear RNAs have a variety of functions, including “splicing” pre-mRNAs and regulating transcription factors.
- 3. UNIT II: GENE EXPRESSION | TRANSCRIPTION IN EUKARYOTES Page 1 of 1 Transcription in Eukaryotes REGULATORY SEQUENCES OF EUKARYOTIC TRANSCRIPTION The regulatory sequences are required for efficient transcription. All these elements bind regulatory proteins (activators and repressors) which help or hinder transcription from the core promoter The regulatory sequences include: 1. Promoters; 2. Enhancers (UAS) 3. Silencers 4. Insulators 1. PROMOTERS Each gene has its own promoter. There may be multiple promoter sequences in a DNA molecule. They indicate transcription start sites. The promoter region lies at the start (or slightly overlaps with) the transcribed region. A promoter of protein coding gene encompasses about 200bp upstream of the transcription initiation site and contains various sequence elements. Promoter can be divided into two regions; a) The Core Promoter Elements b) The Promoter Proximal Elements a) The Core Promoter Elements b)The Promoter Proximal Elements These are Cis-acting sequence needed for the transcription machinery to start the RNA synthesis at the correct site. The elements are typically within 50bps upstream of that site. The best characterized core promoter elements are; i) Initiator (Inr) : A short sequence element which spans the transcription initiation start site. Location between -3 to +5bp Py2CAPy5 A is the first base to be transcribed ii) TATA box or TATA element Goldbery-Hogness box located at about -25bp similar to -10 region of prokaryote. The TATA box has the seven nucleotide consensus sequence. TATAAAA ~15% mammals They align RNA pol at proper site with the help of TFIID The initiator and TATA elements specify where the transcription machinery assembles and determine where transcription will begin. They increase the efficiency of core promoters. They are upstream from the TATA box, in the area from 50 to 200 nucleotides from the start site of transcription. The best characterized promoter proximal elements are; i) CAAT box: It is centered at about -80bp GGCCAATCT ii) GC box: with consensus sequence GGGCGG centered at about -90bp The promoter proximal elements determine how and when a gene is expressed key to this regulation are transcription regulatory proteins called activators, which determine the efficiency of transcription initiation Typically a promoter include 2 or 3 of these core and proximal promoter elements GENERIC PROMOTER
- 4. UNIT II: GENE EXPRESSION | TRANSCRIPTION IN EUKARYOTES Page 1 of 1 Transcription in Eukaryotes THYMIDINE PROMOTER 2. ENHANCERS Enhancers are elements interact with protein other than RNA polymerase and regulate the activity of promoter. They are also called Upstream Activating Sites or UAS. They increase or enhance the efficiency of promoter Location varies; usually 100 or 200 bp upstream which it can be moved several 100s or even 1000 bps upstream or downstream without alteration of their activities. It leads up to 200 fold increase in transcription rate of a gene. 3. SILENCERS These are repressing elements. They can function at great distance from gene (proposed by Alexander Johnson). They turn off the active gene. 4. INSULATORS Insulators block or inhibit or hinder the activity of enhancers. TRANSCRIPTION FACTORS IN EUKARYOTES Unlike the prokaryotic RNA polymerase that can bind to a DNA template on its own, eukaryotes require several other proteins, called transcription factors, to first bind to the promoter region and then help recruit the appropriate polymerase. The completed assembly of transcription factors and RNA polymerase bind to the promoter, forming a transcription pre-initiation complex (PIC). Transcription factors recognize the promoter; RNA polymerase then binds and forms the transcription initiation complex. TYPES OF TRANSCRIPTION FACTORS 1. Basal Transcription factors: Basal transcription factors play a major role in transcription intiation. TFIID, TFIIA, TFIIB, TFIIF, TFIIE, TFIIH, TFIIJ are Basal transcription factors. 2. Upstream Transcription factors: Upstream Transcription factors are also known as activators. They cover upstream region and increase efficiency of initiation. They recognize specific sequences and binds to proximal promoter. SP1, CTF family, CP1 family, C/EBP, ACF, Oct-1, Oct-2, TK
- 5. UNIT II: GENE EXPRESSION | TRANSCRIPTION IN EUKARYOTES Page 1 of 1 Transcription in Eukaryotes 3. Regulatory Transcription factors: Regulatory Transcription factors bind to response elements. They are activated at specific time in specific tissue. BASAL TRANSCRIPTION FACTORS Sl no TRANSCRIPTION FACTORS STRUCTURE FUNCTION 1. TFIID Commitment factor Positioning factor Two sub parts; 1. TATA Binding Protein [TBP] o Highly conserved o binds to minor groove sequence and bends DNA ~800C towards major groove 2. TBP Associating Factor [TAF] binds to major groove and modifies chromatin structure for transcription Multi (12) subunit complex 1 TAF + 11 TBP = 12 Scans DNA Identifies promoter - TATA box Binds to TATA box Initiates transcription 2 TFIIA Multi subunit complex Eg: In yeast two subunits In mammals three subunits Binds to promoter Specifically upstream to TFIID 3 TFIIB Single polypeptide chain Binds to promoter Specifically downstream to TFIID Covers -10 to +10 region Bridges the TATA bound TBP and polymerase 4 TFIIF Two subunits; 1) RAP 74: ATP dependent helicase activity 2) RAP 38: Binds strongly to RNA polymerase Required for the recruitment of RNA Polymerase, TFIIE and TFIIH 5 TFIIE Two subunits Connects RNA polymerase to promoter Downstream +30 bp Recruitment and regulation of TFIIH 6 TFIIH multi subunits (~9) Protein kinase activity (phosphorylates CTD tail of RNA polymerase) ATPase-Helicase activity Promoter melting 7 TFIIJ Undetermined joins TFIIH and TFIIE
- 6. UNIT II: GENE EXPRESSION | TRANSCRIPTION IN EUKARYOTES Page 1 of 1 Transcription in Eukaryotes TRANSCRIPTION STEPS IN EUKARYOTES Accurate initiation of transcription of a protein coding gene involves the assembly of RNA Polymerase II and a number of other proteins called Basal transcription factor on the core promoters. Eukaryotic transcription is carried out in the nucleus of the cell and proceeds in three sequential stages: initiation, elongation, and termination. Transcription involves major three steps; 1) Transcription initiation 2) Transcription elongation 3) Transcription termination 1. TRANSCRIPTION INITIATION Initiation is the first step of eukaryotic transcription and requires RNA polymerase and several transcription factors to proceed. Eukaryotes require transcription factors to first bind to the promoter region and then help recruit the appropriate RNA polymerase. The TATA box, as a core promoter element, is the binding site for a transcription factor known as TATA-binding protein (TBP), which is a subunit of TFIID After TFIID binds to the TATA box via the TBP, five more transcription factors and RNA polymerase combine around the TATA box in a series of stages to form a pre- initiation complex. Transcription Factor II H (TFIIH), is involved in separating opposing strands of double-stranded DNA to provide the RNA Polymerase access to a single-stranded DNA template. The basal transcription factors and RNA Polymerase II bind to promoter elements in a particular order in vivo to produce the complete transcription initiation complex, also called the pre-initiation complex Activators and repressors, along with any associated coactivators or corepressors, are responsible for modulating transcription rate. Activator proteins increase the transcription rate, and repressor proteins decrease the transcription rate.
- 7. UNIT II: GENE EXPRESSION | TRANSCRIPTION IN EUKARYOTES Page 1 of 1 Transcription in Eukaryotes RNA polymerase does not bind to free floating DNA TFIID (TAF + TBP) identifies and binds to TATA box. This permits the association of TFIIA and TFIIB. TFIIA binds upstream TFIIB binds downstream TFIIF + RNA pol II RNA Polymerase is assorted to promoter by TFIIF to form a transcription complex TFII E TFIIH TFIIJ orderly addition of TFIIE, TFIIH and TFIIJ helps in the initiation Formation of pre-initiation complex containing these components is followed by promoter melting which requires hydrolysis of ATP. It is the helicase like activity of TFIIH that stimulates unwinding of promoter. 2. TRANSCRIPTION ELONGATION Once RNA polymerase II has initiated transcription, the elongation phase begins. RNA Polymerase II is a complex of 12 protein subunits. Specific subunits within the protein allow RNA Polymerase II to act as its own helicase, sliding clamp, single- stranded DNA binding protein, as well as carry out other functions. During transition from initiation to elongation, the RNA polymerase II enzyme sheds initiation factors and recruits another set of factors that favor phosphorylation of the CTD in RNA polymerase. Thus, phosphorylation of the CTD leads to an exchange of initiation factors for those factors required for elongation and RNA processing. TFIIH unwinds ds DNA and phosphorylates C-terminal domain of largest subunit of RNA polymerase. RNA polymerase moves along the template strand, synthesising an mRNA molecule, in a same fashion as in prokaryotic transcription. As RNA polymerase accelerates towards downstream, the ribo-nucleotides (A, U, G & C) are added to the template strand, enabling growth of the mRNA transcript. The mRNA transcript is made in a 5’ to 3’ direction, while reading the template DNA strand in the 3' to 5' direction. The template DNA strand and RNA strand are antiparallel.
- 8. UNIT II: GENE EXPRESSION | TRANSCRIPTION IN EUKARYOTES Page 1 of 1 Transcription in Eukaryotes The movement of RNA polymerase is based on the ATP consumption. RNA Polymerases unwind the double stranded DNA ahead of them and allow the unwound DNA behind them to rewind. As a result, RNA strand synthesis occurs in a transcription bubble of about 25 unwound DNA base pairs. Only about 8 nucleotides of newly-synthesized RNA remain base paired to the template DNA. The rest of the RNA molecules fall off the template to allow the DNA behind it to rewind. 3. TRANSCRIPTION TERMINATION In eukaryotes, actual termination of RNA polymearse activity during transcription may take place through termination sites similar to these found in prokaryotes. Yet, the termination of transcription is different for the three different eukaryotic RNA polymerases. These termination sites are believed to be present away from the site of the 3´ end of mRNA are generated due to post transcription cleavage that is achieved by snurp. The sequence 5´-AAUAAA-3´ in mRNA 3´ end seems to be common in eukaryotic mRNA and mutation in these sequence cause elongation of mRNA. The polyadenylation signals act as long terminator regions to end the transcription. RNA Polymerase II has no specific signals that terminate its transcription. In the case of protein-encoding genes, a protein complex will bind to two locations on the growing pre-mRNA once the RNA polymerase has transcribed past the end of the gene. In eukaryotes (such as humans), a primary transcript has to go through some extra processing steps in order to become a mature mRNA. During processing, caps are added to the ends of the RNA, and some pieces of it may be carefully removed in a process called splicing. TERMINATION DIAGRAM – refer class notes – dissolved transcriptosome ( separated pre-mRNA + separated RNA polymerase + transcription factors + ds DNA )
- 9. UNIT II: GENE EXPRESSION | TRANSCRIPTION IN EUKARYOTES Page 1 of 1 Transcription in Eukaryotes Gene expression is the process by which the instructions present in our DNA are converted into a functional product ADDITIONAL INFORMATION In eukaryotes, RNA polymerase II transcribes protein coding genes. The product of transcription is a precursor mRNA or pre-mRNA. The precursor mRNA molecule or transcript that must be modified, processed or both to produce the mature functional mRNA molecule After transcription the RNA molecule is processed in a number of ways: introns are removed and the exons are spliced together to form a mature mRNA molecule consisting of a single protein-coding sequence. They are spliced and have a 5' cap and poly-A tail put on their ends. Capping : Addition of a methylated guanine cap confers protection to the mRNA. This is necessary as RNA is much more unstable than DNA. It involves: Addition of a methylated guanine, Occurs at the 5’ end of the mRNA transcript, Protects the mRNA from degradation Polyadenylation/tailing: Addition of a poly A tail confers protection to the mRNA. This is necessary as RNA is much more unstable than DNA. It involves: Endonucleases* recognise a specific sequence along the mRNA transcript and cleaves it there several adenine nucleotides are added (approximately 200) to the transcript by the enzyme poly A polymerase, Occurs at the 3’ end, protecting the mRNA from degradation. Splicing allows one genetic sequence to code for different proteins. This conserves genetic material. It involves: Introns (non-coding sequences) are removed via spliceosome excision Exons (coding sequence) are then joined together by ligation It is sequence dependent and occurs within (in the middle of) the transcript This allows many proteins to be made from a single pre-mRNA *** BEST WISHES ***
