Bis2C: Lecture 10 extras on "New View of the Tree of Life" paper
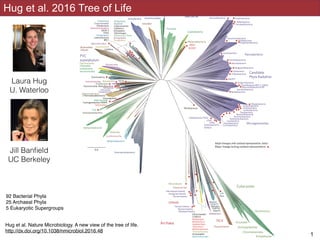
- 1. Hug et al 2016 !1 Hug et al. 2016 Tree of Life 92 Bacterial Phyla 25 Archaeal Phyla 5 Eukaryotic Supergroups Hug et al. Nature Microbiology. A new view of the tree of life. http://dx.doi.org/10.1038/nmicrobiol.2016.48 Laura Hug U. Waterloo Jill Banfield UC Berkeley
- 2. !2 Hug et al 2016Hug et al. 2016 Bacteria Hug et al. Nature Microbiology. A new view of the tree of life. http://dx.doi.org/10.1038/nmicrobiol.2016.48
- 3. Taxa Covered in Textbook !3 Hug et al. Nature Microbiology. A new view of the tree of life. http://dx.doi.org/10.1038/nmicrobiol.2016.48
- 4. !4 Hug et al 2016Phyla Never Grown in the Lab Hug et al. Nature Microbiology. A new view of the tree of life. http://dx.doi.org/10.1038/nmicrobiol.2016.48
- 5. Hug et al 2016 !5 Hug et al. 2016 Archaea and Eukaryotes Hug et al. Nature Microbiology. A new view of the tree of life. http://dx.doi.org/10.1038/nmicrobiol.2016.48
- 6. Hug et al 2016 !6 Hug et al. 2016 Archaea Phyla Never Grown in the Lab Hug et al. Nature Microbiology. A new view of the tree of life. http://dx.doi.org/10.1038/nmicrobiol.2016.48
- 7. Major Groups by Evolutionary Distance !7
- 8. The Dark Matter of Biology !8 0.2 Korarchaeota Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci Archaeoglobi, Methanomicrobia, Halobacteria Aciduliprofundum, Thermoplasmata Uncultured Thermoplasmata Thermoplasmata Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota Cyanobacteria, Melainabacteria Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria Microgenomates Daviesbacteria Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Microgenomates Shapirobacteria Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Microgenomates Gottesmanbacteria KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, KaiserbacteriaParcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi Thermotogae Omnitrophica Omnitrophica Spirochaetes Spirochaetes Hydrogenedentes NKB19 Deltaproteobacteria Epsilonproteobacteria TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria Planctomycetes Chlamydiae Lentisphaerae Verrucomicrobia Verrucomicrobia RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) Aigarchaeota, Cand. Caldiarchaeum subterraneum Unclassified archaea Parcubacteria Candidate Phyla Radiation Cyanobacteria, Melainabacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi q ,q , Thermotogae A ifi C Omnitrophica Omnitrophica pp Spirochaetes Spirochaetes S i h t Hydrogenedentes NKB19 Deltaproteobacteria H d d t N Epsilonproteobacteria b TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria D f bD f b , , p ,, , p , Planctomycetes pp Chlamydiae y Lentisphaerae C a ydCh Verrucomicrobia Verrucomicrobia pp RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) , y g, y g Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria ( ) Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 b M Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) O h Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria gg Microgenomates Daviesbacteria gg Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Mi t L b t i Microgenomates Shapirobacteria Mi t W bMi t Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Mi Sh i b i Microgenomates Gottesmanbacteria t R i b t i g yg y KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria te a Parcubacteria Parcubacteria Parcubacteria Parcubacteria AbscAbs Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, Kaiserbacteria gg Parcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria P b i Candidate Phyla Radiation Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci E43 Archaeoglobi, Methanomicrobia, Halobacteria , , ,, , , Aciduliprofundum, Thermoplasmata gg Uncultured Thermoplasmata p ,p , Thermoplasmata p Unclassified archaea Korarchaeota , Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota b l b Aigarchaeota, FFA Cand. Caldiarchaeum subterraneum C b t i M l i b t i Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Th h , Th h t Eukaryotes Bacteria Archaea Katanobacteria WWE3 Bootstrap ≥ 85% 85% > Bootstrap ≥ 50% Woesearchaeota, Nanoarchaeota gure 2 | A reformatted view of the tree in Fig. 1 in which each major lineage represents the same amount of evolutionary distance. The threshold for oups (coloured wedges) was an average branch length of <0.65 substitutions per site. Notably, some well-accepted phyla become single groups and hers are split into multiple distinct groups. We undertook this analysis to provide perspective on the structure of the tree, and do not propose the resulting oups to have special taxonomic status. The massive scale of diversity in the CPR and the large fraction of major lineages that lack isolated representatives ed dots) are apparent from this analysis. Bootstrap support values are indicated by circles on nodes—black for support of 85% and above, grey for support m 50 to 84%. The complete ribosomal protein tree is available in rectangular format with full bootstrap values as Supplementary Fig. 1 and in Newick mat in Supplementary Dataset 2. ATURE MICROBIOLOGY DOI: 10.1038/NMICROBIOL.2016.48 LETTERS ATURE MICROBIOLOGY | www.nature.com/naturemicrobiology 3 © 2016 Macmillan Publishers Limited. All rights reserved 2 Korarchaeota Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci Archaeoglobi, Methanomicrobia, Halobacteria Aciduliprofundum, Thermoplasmata Uncultured Thermoplasmata Thermoplasmata Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota Cyanobacteria, Melainabacteria Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria Microgenomates Daviesbacteria Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Microgenomates Shapirobacteria Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Microgenomates Gottesmanbacteria KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, KaiserbacteriaParcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi Thermotogae Omnitrophica Omnitrophica Spirochaetes Spirochaetes Hydrogenedentes NKB19 Deltaproteobacteria Epsilonproteobacteria TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria Planctomycetes Chlamydiae Lentisphaerae Verrucomicrobia Verrucomicrobia RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) Aigarchaeota, Cand. Caldiarchaeum subterraneum Unclassified archaea Parcubacteria Candidate Phyla Radiation Cyanobacteria, Melainabacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi q ,q , Thermotogae A ifi C Omnitrophica Omnitrophica pp Spirochaetes Spirochaetes S i h t Hydrogenedentes NKB19 Deltaproteobacteria H d d t N Epsilonproteobacteria b TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria D f bD f b , , p ,, , p , Planctomycetes pp Chlamydiae y Lentisphaerae C a ydCh Verrucomicrobia Verrucomicrobia pp RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) , y g, y g Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria ( ) Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 b M Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) O h Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria gg Microgenomates Daviesbacteria gg Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Mi t L b t i Microgenomates Shapirobacteria Mi t W bMi t Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Mi Sh i b i Microgenomates Gottesmanbacteria t R i b t i g yg y KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria te a Parcubacteria Parcubacteria Parcubacteria Parcubacteria AbscAbs Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, Kaiserbacteria gg Parcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria P b i Candidate Phyla Radiation Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci E43 Archaeoglobi, Methanomicrobia, Halobacteria , , ,, , , Aciduliprofundum, Thermoplasmata gg Uncultured Thermoplasmata p ,p , Thermoplasmata p Unclassified archaea Korarchaeota , Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota b l b Aigarchaeota, FFA Cand. Caldiarchaeum subterraneum C b t i M l i b t i Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Th h , Th h t Eukaryotes Bacteria Archaea Katanobacteria WWE3 Bootstrap ≥ 85% 85% > Bootstrap ≥ 50% Woesearchaeota, Nanoarchaeota A reformatted view of the tree in Fig. 1 in which each major lineage represents the same amount of evolutionary distance. The threshold for oured wedges) was an average branch length of <0.65 substitutions per site. Notably, some well-accepted phyla become single groups and plit into multiple distinct groups. We undertook this analysis to provide perspective on the structure of the tree, and do not propose the resulting ave special taxonomic status. The massive scale of diversity in the CPR and the large fraction of major lineages that lack isolated representatives re apparent from this analysis. Bootstrap support values are indicated by circles on nodes—black for support of 85% and above, grey for support 84%. The complete ribosomal protein tree is available in rectangular format with full bootstrap values as Supplementary Fig. 1 and in Newick upplementary Dataset 2. E MICROBIOLOGY DOI: 10.1038/NMICROBIOL.2016.48 LETTERS ROBIOLOGY | www.nature.com/naturemicrobiology 3 © 2016 Macmillan Publishers Limited. All rights reserved 0.2 Korarchaeota Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci Archaeoglobi, Methanomicrobia, Halobacteria Aciduliprofundum, Thermoplasmata Uncultured Thermoplasmata Thermoplasmata Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota Cyanobacteria, Melainabacteria Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria Microgenomates Daviesbacteria Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Microgenomates Shapirobacteria Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Microgenomates Gottesmanbacteria KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, KaiserbacteriaParcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi Thermotogae Omnitrophica Omnitrophica Spirochaetes Spirochaetes Hydrogenedentes NKB19 Deltaproteobacteria Epsilonproteobacteria TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria Planctomycetes Chlamydiae Lentisphaerae Verrucomicrobia Verrucomicrobia RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) Aigarchaeota, Cand. Caldiarchaeum subterraneum Unclassified archaea Parcubacteria Candidate Phyla Radiation Cyanobacteria, Melainabacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi q ,q , Thermotogae A ifi C Omnitrophica Omnitrophica pp Spirochaetes Spirochaetes S i h t Hydrogenedentes NKB19 Deltaproteobacteria H d d t N Epsilonproteobacteria b TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria D f bD f b , , p ,, , p , Planctomycetes pp Chlamydiae y Lentisphaerae C a ydCh Verrucomicrobia Verrucomicrobia pp RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) , y g, y g Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria ( ) Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 b M Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) O h Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria gg Microgenomates Daviesbacteria gg Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Mi t L b t i Microgenomates Shapirobacteria Mi t W bMi t Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Mi Sh i b i Microgenomates Gottesmanbacteria t R i b t i g yg y KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria te a Parcubacteria Parcubacteria Parcubacteria Parcubacteria AbscAbs Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, Kaiserbacteria gg Parcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria P b i Candidate Phyla Radiation Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci E43 Archaeoglobi, Methanomicrobia, Halobacteria , , ,, , , Aciduliprofundum, Thermoplasmata gg Uncultured Thermoplasmata p ,p , Thermoplasmata p Unclassified archaea Korarchaeota , Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota b l b Aigarchaeota, FFA Cand. Caldiarchaeum subterraneum C b t i M l i b t i Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Th h , Th h t Eukaryotes Bacteria Archaea Katanobacteria WWE3 Bootstrap ≥ 85% 85% > Bootstrap ≥ 50% Woesearchaeota, Nanoarchaeota Figure 2 | A reformatted view of the tree in Fig. 1 in which each major lineage represents the same amount of evolutionary distance. The threshold for groups (coloured wedges) was an average branch length of <0.65 substitutions per site. Notably, some well-accepted phyla become single groups and others are split into multiple distinct groups. We undertook this analysis to provide perspective on the structure of the tree, and do not propose the resulting groups to have special taxonomic status. The massive scale of diversity in the CPR and the large fraction of major lineages that lack isolated representatives (red dots) are apparent from this analysis. Bootstrap support values are indicated by circles on nodes—black for support of 85% and above, grey for support from 50 to 84%. The complete ribosomal protein tree is available in rectangular format with full bootstrap values as Supplementary Fig. 1 and in Newick format in Supplementary Dataset 2. NATURE MICROBIOLOGY DOI: 10.1038/NMICROBIOL.2016.48 LETTERS NATURE MICROBIOLOGY | www.nature.com/naturemicrobiology 3 © 2016 Macmillan Publishers Limited. All rights reserved 0.2 Korarchaeota Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci Archaeoglobi, Methanomicrobia, Halobacteria Aciduliprofundum, Thermoplasmata Uncultured Thermoplasmata Thermoplasmata Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota Cyanobacteria, Melainabacteria Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria Microgenomates Daviesbacteria Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Microgenomates Shapirobacteria Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Microgenomates Gottesmanbacteria KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, KaiserbacteriaParcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi Thermotogae Omnitrophica Omnitrophica Spirochaetes Spirochaetes Hydrogenedentes NKB19 Deltaproteobacteria Epsilonproteobacteria TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria Planctomycetes Chlamydiae Lentisphaerae Verrucomicrobia Verrucomicrobia RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) Aigarchaeota, Cand. Caldiarchaeum subterraneum Unclassified archaea Parcubacteria Candidate Phyla Radiation Cyanobacteria, Melainabacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi q ,q , Thermotogae A ifi C Omnitrophica Omnitrophica pp Spirochaetes Spirochaetes S i h t Hydrogenedentes NKB19 Deltaproteobacteria H d d t N Epsilonproteobacteria b TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria D f bD f b , , p ,, , p , Planctomycetes pp Chlamydiae y Lentisphaerae C a ydCh Verrucomicrobia Verrucomicrobia pp RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) , y g, y g Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria ( ) Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 b M Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) O h Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria gg Microgenomates Daviesbacteria gg Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Mi t L b t i Microgenomates Shapirobacteria Mi t W bMi t Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Mi Sh i b i Microgenomates Gottesmanbacteria t R i b t i g yg y KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria te a Parcubacteria Parcubacteria Parcubacteria Parcubacteria AbscAbs Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, Kaiserbacteria gg Parcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria P b i Candidate Phyla Radiation Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci E43 Archaeoglobi, Methanomicrobia, Halobacteria , , ,, , , Aciduliprofundum, Thermoplasmata gg Uncultured Thermoplasmata p ,p , Thermoplasmata p Unclassified archaea Korarchaeota , Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota b l b Aigarchaeota, FFA Cand. Caldiarchaeum subterraneum C b t i M l i b t i Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Th h , Th h t Eukaryotes Bacteria Archaea Katanobacteria WWE3 Bootstrap ≥ 85% 85% > Bootstrap ≥ 50% Woesearchaeota, Nanoarchaeota Figure 2 | A reformatted view of the tree in Fig. 1 in which each major lineage represents the same amount of evolutionary distance. The threshold for groups (coloured wedges) was an average branch length of <0.65 substitutions per site. Notably, some well-accepted phyla become single groups and others are split into multiple distinct groups. We undertook this analysis to provide perspective on the structure of the tree, and do not propose the resulting groups to have special taxonomic status. The massive scale of diversity in the CPR and the large fraction of major lineages that lack isolated representatives (red dots) are apparent from this analysis. Bootstrap support values are indicated by circles on nodes—black for support of 85% and above, grey for support from 50 to 84%. The complete ribosomal protein tree is available in rectangular format with full bootstrap values as Supplementary Fig. 1 and in Newick format in Supplementary Dataset 2. NATURE MICROBIOLOGY DOI: 10.1038/NMICROBIOL.2016.48 LETTERS NATURE MICROBIOLOGY | www.nature.com/naturemicrobiology 3 © 2016 Macmillan Publishers Limited. All rights reserved Korarchaeota Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci Archaeoglobi, Methanomicrobia, Halobacteria Aciduliprofundum, Thermoplasmata Uncultured Thermoplasmata Thermoplasmata Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota Cyanobacteria, Melainabacteria Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria Microgenomates Daviesbacteria Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Microgenomates Shapirobacteria Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Microgenomates Gottesmanbacteria KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, KaiserbacteriaParcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi Thermotogae Omnitrophica Omnitrophica Spirochaetes Spirochaetes Hydrogenedentes NKB19 Deltaproteobacteria Epsilonproteobacteria TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria Planctomycetes Chlamydiae Lentisphaerae Verrucomicrobia Verrucomicrobia RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) Aigarchaeota, Cand. Caldiarchaeum subterraneum Unclassified archaea Parcubacteria Candidate Phyla Radiation Cyanobacteria, Melainabacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi q ,q , Thermotogae A ifi C Omnitrophica Omnitrophica pp Spirochaetes Spirochaetes S i h t Hydrogenedentes NKB19 Deltaproteobacteria H d d t N Epsilonproteobacteria b TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria D f bD f b , , p ,, , p , Planctomycetes pp Chlamydiae y Lentisphaerae C a ydCh Verrucomicrobia Verrucomicrobia pp RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) , y g, y g Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria ( ) Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 b M Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) O h Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria gg Microgenomates Daviesbacteria gg Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Mi t L b t i Microgenomates Shapirobacteria Mi t W bMi t Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Mi Sh i b i Microgenomates Gottesmanbacteria t R i b t i g yg y KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria te a Parcubacteria Parcubacteria Parcubacteria Parcubacteria AbscAbs Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, Kaiserbacteria gg Parcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria P b i Candidate Phyla Radiation Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci E43 Archaeoglobi, Methanomicrobia, Halobacteria , , ,, , , Aciduliprofundum, Thermoplasmata gg Uncultured Thermoplasmata p ,p , Thermoplasmata p Unclassified archaea Korarchaeota , Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota b l b Aigarchaeota, FFA Cand. Caldiarchaeum subterraneum C b t i M l i b t i Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Th h , Th h t Eukaryotes Bacteria Archaea Katanobacteria WWE3 Bootstrap ≥ 85% 85% > Bootstrap ≥ 50% Woesearchaeota, Nanoarchaeota matted view of the tree in Fig. 1 in which each major lineage represents the same amount of evolutionary distance. The threshold for edges) was an average branch length of <0.65 substitutions per site. Notably, some well-accepted phyla become single groups and multiple distinct groups. We undertook this analysis to provide perspective on the structure of the tree, and do not propose the resulting cial taxonomic status. The massive scale of diversity in the CPR and the large fraction of major lineages that lack isolated representatives rent from this analysis. Bootstrap support values are indicated by circles on nodes—black for support of 85% and above, grey for support he complete ribosomal protein tree is available in rectangular format with full bootstrap values as Supplementary Fig. 1 and in Newick ntary Dataset 2. CROBIOLOGY DOI: 10.1038/NMICROBIOL.2016.48 LETTERS 0.2 Korarchaeota Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci Archaeoglobi, Methanomicrobia, Halobacteria Aciduliprofundum, Thermoplasmata Uncultured Thermoplasmata Thermoplasmata Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota Cyanobacteria, Melainabacteria Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria Microgenomates Daviesbacteria Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Microgenomates Shapirobacteria Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Microgenomates Gottesmanbacteria KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, KaiserbacteriaParcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi Thermotogae Omnitrophica Omnitrophica Spirochaetes Spirochaetes Hydrogenedentes NKB19 Deltaproteobacteria Epsilonproteobacteria TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria Planctomycetes Chlamydiae Lentisphaerae Verrucomicrobia Verrucomicrobia RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) Aigarchaeota, Cand. Caldiarchaeum subterraneum Unclassified archaea Parcubacteria Candidate Phyla Radiation Cyanobacteria, Melainabacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi q ,q , Thermotogae A ifi C Omnitrophica Omnitrophica pp Spirochaetes Spirochaetes S i h t Hydrogenedentes NKB19 Deltaproteobacteria H d d t N Epsilonproteobacteria b TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria D f bD f b , , p ,, , p , Planctomycetes pp Chlamydiae y Lentisphaerae C a ydCh Verrucomicrobia Verrucomicrobia pp RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) , y g, y g Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria ( ) Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 b M Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) O h Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria gg Microgenomates Daviesbacteria gg Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Mi t L b t i Microgenomates Shapirobacteria Mi t W bMi t Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Mi Sh i b i Microgenomates Gottesmanbacteria t R i b t i g yg y KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria te a Parcubacteria Parcubacteria Parcubacteria Parcubacteria AbscAbs Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, Kaiserbacteria gg Parcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria P b i Candidate Phyla Radiation Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci E43 Archaeoglobi, Methanomicrobia, Halobacteria , , ,, , , Aciduliprofundum, Thermoplasmata gg Uncultured Thermoplasmata p ,p , Thermoplasmata p Unclassified archaea Korarchaeota , Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota b l b Aigarchaeota, FFA Cand. Caldiarchaeum subterraneum C b t i M l i b t i Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Th h , Th h t Eukaryotes Bacteria Archaea Katanobacteria WWE3 Bootstrap ≥ 85% 85% > Bootstrap ≥ 50% Woesearchaeota, Nanoarchaeota Figure 2 | A reformatted view of the tree in Fig. 1 in which each major lineage represents the same amount of evolutionary distance. The threshold for groups (coloured wedges) was an average branch length of <0.65 substitutions per site. Notably, some well-accepted phyla become single groups and others are split into multiple distinct groups. We undertook this analysis to provide perspective on the structure of the tree, and do not propose the resulting groups to have special taxonomic status. The massive scale of diversity in the CPR and the large fraction of major lineages that lack isolated representatives (red dots) are apparent from this analysis. Bootstrap support values are indicated by circles on nodes—black for support of 85% and above, grey for support from 50 to 84%. The complete ribosomal protein tree is available in rectangular format with full bootstrap values as Supplementary Fig. 1 and in Newick format in Supplementary Dataset 2. NATURE MICROBIOLOGY DOI: 10.1038/NMICROBIOL.2016.48 LETTERS NATURE MICROBIOLOGY | www.nature.com/naturemicrobiology 3 © 2016 Macmillan Publishers Limited. All rights reserved 0.2 Korarchaeota Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci Archaeoglobi, Methanomicrobia, Halobacteria Aciduliprofundum, Thermoplasmata Uncultured Thermoplasmata Thermoplasmata Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota Cyanobacteria, Melainabacteria Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria Microgenomates Daviesbacteria Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Microgenomates Shapirobacteria Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Microgenomates Gottesmanbacteria KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, KaiserbacteriaParcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi Thermotogae Omnitrophica Omnitrophica Spirochaetes Spirochaetes Hydrogenedentes NKB19 Deltaproteobacteria Epsilonproteobacteria TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria Planctomycetes Chlamydiae Lentisphaerae Verrucomicrobia Verrucomicrobia RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) Aigarchaeota, Cand. Caldiarchaeum subterraneum Unclassified archaea Parcubacteria Candidate Phyla Radiation Cyanobacteria, Melainabacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi q ,q , Thermotogae A ifi C Omnitrophica Omnitrophica pp Spirochaetes Spirochaetes S i h t Hydrogenedentes NKB19 Deltaproteobacteria H d d t N Epsilonproteobacteria b TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria D f bD f b , , p ,, , p , Planctomycetes pp Chlamydiae y Lentisphaerae C a ydCh Verrucomicrobia Verrucomicrobia pp RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) , y g, y g Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria ( ) Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 b M Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) O h Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria gg Microgenomates Daviesbacteria gg Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Mi t L b t i Microgenomates Shapirobacteria Mi t W bMi t Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Mi Sh i b i Microgenomates Gottesmanbacteria t R i b t i g yg y KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria te a Parcubacteria Parcubacteria Parcubacteria Parcubacteria AbscAbs Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, Kaiserbacteria gg Parcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria P b i Candidate Phyla Radiation Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci E43 Archaeoglobi, Methanomicrobia, Halobacteria , , ,, , , Aciduliprofundum, Thermoplasmata gg Uncultured Thermoplasmata p ,p , Thermoplasmata p Unclassified archaea Korarchaeota , Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota b l b Aigarchaeota, FFA Cand. Caldiarchaeum subterraneum C b t i M l i b t i Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Th h , Th h t Eukaryotes Bacteria Archaea Katanobacteria WWE3 Bootstrap ≥ 85% 85% > Bootstrap ≥ 50% Woesearchaeota, Nanoarchaeota Figure 2 | A reformatted view of the tree in Fig. 1 in which each major lineage represents the same amount of evolutionary distance. The threshold for groups (coloured wedges) was an average branch length of <0.65 substitutions per site. Notably, some well-accepted phyla become single groups and others are split into multiple distinct groups. We undertook this analysis to provide perspective on the structure of the tree, and do not propose the resulting groups to have special taxonomic status. The massive scale of diversity in the CPR and the large fraction of major lineages that lack isolated representatives (red dots) are apparent from this analysis. Bootstrap support values are indicated by circles on nodes—black for support of 85% and above, grey for support from 50 to 84%. The complete ribosomal protein tree is available in rectangular format with full bootstrap values as Supplementary Fig. 1 and in Newick format in Supplementary Dataset 2. NATURE MICROBIOLOGY DOI: 10.1038/NMICROBIOL.2016.48 LETTERS NATURE MICROBIOLOGY | www.nature.com/naturemicrobiology 3 © 2016 Macmillan Publishers Limited. All rights reserved cteria esebacteria cteria bacteria,Pacebacteria,Collierbacteria eria CandidatePhylaRadiation cteria esebacteria cteria b bacteria,Pacebacteria,Collierbacteria i eria CandidatePhylaRadiation cteria esebacteria cteria bacteria,Pacebacteria,Collierbacteria eria CandidatePhylaRadiation cteria esebacteria cteria b bacteria,Pacebacteria,Collierbacteria i eria CandidatePhylaRadiation Korarchaeota Woesearchaeota,Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri,Methanococci,Methanobacteria,Hadesarchaea,Thermococci Archaeoglobi,Methanomicrobia,Halobacteria Aciduliprofundum,Thermoplasmata UnculturedThermoplasmata Thermoplasmata Opisthokonta,Excavata,Archaeplastida Chromalveolata,Amoebozoa Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota Cyanobacteria,Melainabacteria Deinococcus-Thermus Aquificae,CalescamantesEM19 Caldiserica,Dictyoglomi Thermotogae RBX-1 WOR-1 Firmicutes,Tenericutes,Armatimonadetes,Chloroflexi,Actinobacteria Fusobacteria,Synergistetes Unculturedbacteria(CPRIF32) bacteria(OP9) BRC1,Poribacteria Aigarchaeota,Cand.Caldiarchaeumsubterraneum Cyanobacteria,Melainabacteria Deinococcus-Thermus Aquificae,CalescamantesEM19 Caldiserica,Dictyoglomi q ,q , Thermotogae A ifi C RBX-1 WOR-1 Firmicutes,Tenericutes,Armatimonadetes,Chloroflexi,Actinobacteria Fusobacteria,Synergistetes Unculturedbacteria(CPRIF32) , y g, y g bacteria(OP9) BRC1,Poribacteria ( ) Woesearchaeota,Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri,Methanococci,Methanobacteria,Hadesarchaea,Thermococci E43 Archaeoglobi,Methanomicrobia,Halobacteria , , ,, , , Aciduliprofundum,Thermoplasmata gg UnculturedThermoplasmata p ,p , Thermoplasmata p Korarchaeota , Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota b l b Aigarchaeota, FFA Cand.Caldiarchaeumsubterraneum C b t i M l i b t i Opisthokonta,Excavata,Archaeplastida Chromalveolata,Amoebozoa Th h , Th h t Eukaryotes Bacteria ArchaeaWoesearchaeota,Nanoarchaeota Korarchaeota Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci Archaeoglobi, Methanomicrobia, Halobacteria Aciduliprofundum, Thermoplasmata Uncultured Thermoplasmata Thermoplasmata Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota Cyanobacteria, Melainabacteria Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria Microgenomates Daviesbacteria Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Microgenomates Shapirobacteria Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Microgenomates Gottesmanbacteria KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, KaiserbacteriaParcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi Thermotogae Omnitrophica Omnitrophica Spirochaetes Spirochaetes Hydrogenedentes NKB19 Deltaproteobacteria Epsilonproteobacteria TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria Planctomycetes Chlamydiae Lentisphaerae Verrucomicrobia Verrucomicrobia RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) Aigarchaeota, Cand. Caldiarchaeum subterraneum Unclassified archaea Parcubacteria Candidate Phyla Radiation Cyanobacteria, Melainabacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi q ,q , Thermotogae A ifi C Omnitrophica Omnitrophica pp Spirochaetes Spirochaetes S i h t Hydrogenedentes NKB19 Deltaproteobacteria H d d t N Epsilonproteobacteria b TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria D f bD f b , , p ,, , p , Planctomycetes pp Chlamydiae y Lentisphaerae C a ydCh Verrucomicrobia Verrucomicrobia pp RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) , y g, y g Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria ( ) Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 b M Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) O h Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria gg Microgenomates Daviesbacteria gg Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Mi t L b t i Microgenomates Shapirobacteria Mi t W bMi t Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Mi Sh i b i Microgenomates Gottesmanbacteria t R i b t i g yg y KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria te a Parcubacteria Parcubacteria Parcubacteria Parcubacteria AbscAbs Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, Kaiserbacteria gg Parcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria P b i Candidate Phyla Radiation Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci E43 Archaeoglobi, Methanomicrobia, Halobacteria , , ,, , , Aciduliprofundum, Thermoplasmata gg Uncultured Thermoplasmata p ,p , Thermoplasmata p Unclassified archaea Korarchaeota , Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota b l b Aigarchaeota, FFA Cand. Caldiarchaeum subterraneum C b t i M l i b t i Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Th h , Th h t Eukaryotes Bacteria Archaea Katanobacteria WWE3 Bootstrap ≥ 85% 85% > Bootstrap ≥ 50% Woesearchaeota, Nanoarchaeota of the tree in Fig. 1 in which each major lineage represents the same amount of evolutionary distance. The threshold for an average branch length of <0.65 substitutions per site. Notably, some well-accepted phyla become single groups and tinct groups. We undertook this analysis to provide perspective on the structure of the tree, and do not propose the resulting ic status. The massive scale of diversity in the CPR and the large fraction of major lineages that lack isolated representatives is analysis. Bootstrap support values are indicated by circles on nodes—black for support of 85% and above, grey for support ribosomal protein tree is available in rectangular format with full bootstrap values as Supplementary Fig. 1 and in Newick et 2. OLOGY DOI: 10.1038/NMICROBIOL.2016.48 LETTERS ature.com/naturemicrobiology 3 © 2016 Macmillan Publishers Limited. All rights reserved Korarchaeota Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci Archaeoglobi, Methanomicrobia, Halobacteria Aciduliprofundum, Thermoplasmata Uncultured Thermoplasmata Thermoplasmata Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota Cyanobacteria, Melainabacteria Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria Microgenomates Daviesbacteria Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Microgenomates Shapirobacteria Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Microgenomates Gottesmanbacteria KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, KaiserbacteriaParcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi Thermotogae Omnitrophica Omnitrophica Spirochaetes Spirochaetes Hydrogenedentes NKB19 Deltaproteobacteria Epsilonproteobacteria TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria Planctomycetes Chlamydiae Lentisphaerae Verrucomicrobia Verrucomicrobia RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) Aigarchaeota, Cand. Caldiarchaeum subterraneum Unclassified archaea Parcubacteria Candidate Phyla Radiation Cyanobacteria, Melainabacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi q ,q , Thermotogae A ifi C Omnitrophica Omnitrophica pp Spirochaetes Spirochaetes S i h t Hydrogenedentes NKB19 Deltaproteobacteria H d d t N Epsilonproteobacteria b TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria D f bD f b , , p ,, , p , Planctomycetes pp Chlamydiae y Lentisphaerae C a ydCh Verrucomicrobia Verrucomicrobia pp RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) , y g, y g Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria ( ) Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 b M Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) O h Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria gg Microgenomates Daviesbacteria gg Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Mi t L b t i Microgenomates Shapirobacteria Mi t W bMi t Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Mi Sh i b i Microgenomates Gottesmanbacteria t R i b t i g yg y KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria te a Parcubacteria Parcubacteria Parcubacteria Parcubacteria AbscAbs Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, Kaiserbacteria gg Parcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria P b i Candidate Phyla Radiation Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci E43 Archaeoglobi, Methanomicrobia, Halobacteria , , ,, , , Aciduliprofundum, Thermoplasmata gg Uncultured Thermoplasmata p ,p , Thermoplasmata p Unclassified archaea Korarchaeota , Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota b l b Aigarchaeota, FFA Cand. Caldiarchaeum subterraneum C b t i M l i b t i Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Th h , Th h t Eukaryotes Bacteria Archaea Katanobacteria WWE3 Bootstrap ≥ 85% 85% > Bootstrap ≥ 50% Woesearchaeota, Nanoarchaeota view of the tree in Fig. 1 in which each major lineage represents the same amount of evolutionary distance. The threshold for ) was an average branch length of <0.65 substitutions per site. Notably, some well-accepted phyla become single groups and ple distinct groups. We undertook this analysis to provide perspective on the structure of the tree, and do not propose the resulting xonomic status. The massive scale of diversity in the CPR and the large fraction of major lineages that lack isolated representatives rom this analysis. Bootstrap support values are indicated by circles on nodes—black for support of 85% and above, grey for support mplete ribosomal protein tree is available in rectangular format with full bootstrap values as Supplementary Fig. 1 and in Newick Dataset 2. OBIOLOGY DOI: 10.1038/NMICROBIOL.2016.48 LETTERS www.nature.com/naturemicrobiology 3 © 2016 Macmillan Publishers Limited. All rights reserved Korarchaeota Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci Archaeoglobi, Methanomicrobia, Halobacteria Aciduliprofundum, Thermoplasmata Uncultured Thermoplasmata Thermoplasmata Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota Cyanobacteria, Melainabacteria Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria Microgenomates Daviesbacteria Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Microgenomates Shapirobacteria Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Microgenomates Gottesmanbacteria KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, KaiserbacteriaParcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi Thermotogae Omnitrophica Omnitrophica Spirochaetes Spirochaetes Hydrogenedentes NKB19 Deltaproteobacteria Epsilonproteobacteria TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria Planctomycetes Chlamydiae Lentisphaerae Verrucomicrobia Verrucomicrobia RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) Aigarchaeota, Cand. Caldiarchaeum subterraneum Unclassified archaea Parcubacteria Candidate Phyla Radiation Cyanobacteria, Melainabacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi q ,q , Thermotogae A ifi C Omnitrophica Omnitrophica pp Spirochaetes Spirochaetes S i h t Hydrogenedentes NKB19 Deltaproteobacteria H d d t N Epsilonproteobacteria b TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria D f bD f b , , p ,, , p , Planctomycetes pp Chlamydiae y Lentisphaerae C a ydCh Verrucomicrobia Verrucomicrobia pp RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) , y g, y g Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria ( ) Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 b M Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) O h Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria gg Microgenomates Daviesbacteria gg Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Mi t L b t i Microgenomates Shapirobacteria Mi t W bMi t Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Mi Sh i b i Microgenomates Gottesmanbacteria t R i b t i g yg y KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria te a Parcubacteria Parcubacteria Parcubacteria Parcubacteria AbscAbs Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, Kaiserbacteria gg Parcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria P b i Candidate Phyla Radiation Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci E43 Archaeoglobi, Methanomicrobia, Halobacteria , , ,, , , Aciduliprofundum, Thermoplasmata gg Uncultured Thermoplasmata p ,p , Thermoplasmata p Unclassified archaea Korarchaeota , Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota b l b Aigarchaeota, FFA Cand. Caldiarchaeum subterraneum C b t i M l i b t i Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Th h , Th h t Eukaryotes Bacteria Archaea Katanobacteria WWE3 Bootstrap ≥ 85% 85% > Bootstrap ≥ 50% Woesearchaeota, Nanoarchaeota e tree in Fig. 1 in which each major lineage represents the same amount of evolutionary distance. The threshold for erage branch length of <0.65 substitutions per site. Notably, some well-accepted phyla become single groups and groups. We undertook this analysis to provide perspective on the structure of the tree, and do not propose the resulting atus. The massive scale of diversity in the CPR and the large fraction of major lineages that lack isolated representatives alysis. Bootstrap support values are indicated by circles on nodes—black for support of 85% and above, grey for support omal protein tree is available in rectangular format with full bootstrap values as Supplementary Fig. 1 and in Newick OGY DOI: 10.1038/NMICROBIOL.2016.48 LETTERS om/naturemicrobiology 3 © 2016 Macmillan Publishers Limited. All rights reserved Korarchaeota Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci Archaeoglobi, Methanomicrobia, Halobacteria Aciduliprofundum, Thermoplasmata Uncultured Thermoplasmata Thermoplasmata Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota Cyanobacteria, Melainabacteria Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria Microgenomates Daviesbacteria Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Microgenomates Shapirobacteria Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Microgenomates Gottesmanbacteria KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, KaiserbacteriaParcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi Thermotogae Omnitrophica Omnitrophica Spirochaetes Spirochaetes Hydrogenedentes NKB19 Deltaproteobacteria Epsilonproteobacteria TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria Planctomycetes Chlamydiae Lentisphaerae Verrucomicrobia Verrucomicrobia RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) Aigarchaeota, Cand. Caldiarchaeum subterraneum Unclassified archaea Parcubacteria Candidate Phyla Radiation Cyanobacteria, Melainabacteria Deinococcus-Thermus Aquificae, Calescamantes EM19 Caldiserica, Dictyoglomi q ,q , Thermotogae A ifi C Omnitrophica Omnitrophica pp Spirochaetes Spirochaetes S i h t Hydrogenedentes NKB19 Deltaproteobacteria H d d t N Epsilonproteobacteria b TM6 Alphaproteobacteria, Zetaproteobacteria, Betaproteobacteria, GammaproteobacteriaChrysiogenetes, Deferribacteres Modulibacteria, Tectomicrobia, Nitrospinae, Nitrospirae, Dadabacteria, Thermodesulfobacteria, Deltaprot. NC10, Rokubacteria, Aminicenantes, Acidobacteria D f bD f b , , p ,, , p , Planctomycetes pp Chlamydiae y Lentisphaerae C a ydCh Verrucomicrobia Verrucomicrobia pp RBX-1 WOR-1 Firmicutes, Tenericutes, Armatimonadetes, Chloroflexi, Actinobacteria Fusobacteria, Synergistetes Uncultured bacteria (CP RIF32) , y g, y g Zixibacteria, Marinimicrobia, Caldithrix, Chlorobi, Ignavibacteria, Bacteroidetes Fibrobacteres Cloacamonetes Atribacteria (OP9) BRC1, Poribacteria ( ) Latescibacteria WS3 Gemmatimonadetes, WOR-3, TA06 b M Elusimicrobia Uncultured bacteria Uncultured bacteria (CP RIF1) O h Dojkabacteria WS6 CPR3 Katanobacteria WWE3 Katanobacteria WWE3 Microgenomates Roizmanbacteria Microgenomates Roizmanbacteria Microgenomates Microgenomates Curtissbacteria gg Microgenomates Daviesbacteria gg Microgenomates Levybacteria Microgenomates Woesebacteria Microgenomates Amesbacteria Mi t L b t i Microgenomates Shapirobacteria Mi t W bMi t Microgenomates Beckwithbacteria, Pacebacteria, Collierbacteria Mi Sh i b i Microgenomates Gottesmanbacteria t R i b t i g yg y KAZAN CPR2, Saccharibacteria TM7 Berkelbacteria Berkelbacteria Berkelbacteria Berkelbacteria CPR Uncultured unclassified bacteria Peregrinibacteria Peregrinibacteria Absconditabacteria SR1 Gracilibacteria BD1-5 / GNO2 SM2F11 Parcubacteria Parcubacteria Kuenenbacteria, Falkowbacteria, Uhrbacteria, Magasanikbacteria te a Parcubacteria Parcubacteria Parcubacteria Parcubacteria AbscAbs Parcubacteria Azambacteria, Jorgensenbacteria, Wolfebacteria, Giovannonibacteria, Nomurabacteria, Campbellbacteria, Adlerbacteria, Kaiserbacteria gg Parcubacteria Parcubacteria Moranbacteria Parcubacteria Parcubacteria Yanofskybacteria P b i Candidate Phyla Radiation Diapherotrites Nanohaloarchaeota Unclassified archaea Pacearchaeota Woesearchaeota, Nanoarchaeota Woesearchaeota Altiarchaeales Z7ME43 Methanopyri, Methanococci, Methanobacteria, Hadesarchaea, Thermococci E43 Archaeoglobi, Methanomicrobia, Halobacteria , , ,, , , Aciduliprofundum, Thermoplasmata gg Uncultured Thermoplasmata p ,p , Thermoplasmata p Unclassified archaea Korarchaeota , Crenarchaeota Crenarchaeota Thorarchaeota Lokiarchaeota YNPFFA Thaumarchaeota Thaumarchaeota b l b Aigarchaeota, FFA Cand. Caldiarchaeum subterraneum C b t i M l i b t i Opisthokonta, Excavata, Archaeplastida Chromalveolata, Amoebozoa Th h , Th h t Eukaryotes Bacteria Archaea Katanobacteria WWE3 Bootstrap ≥ 85% 85% > Bootstrap ≥ 50% Woesearchaeota, Nanoarchaeota view of the tree in Fig. 1 in which each major lineage represents the same amount of evolutionary distance. The threshold for ) was an average branch length of <0.65 substitutions per site. Notably, some well-accepted phyla become single groups and ple distinct groups. We undertook this analysis to provide perspective on the structure of the tree, and do not propose the resulting xonomic status. The massive scale of diversity in the CPR and the large fraction of major lineages that lack isolated representatives rom this analysis. Bootstrap support values are indicated by circles on nodes—black for support of 85% and above, grey for support mplete ribosomal protein tree is available in rectangular format with full bootstrap values as Supplementary Fig. 1 and in Newick Dataset 2. OBIOLOGY DOI: 10.1038/NMICROBIOL.2016.48 LETTERS www.nature.com/naturemicrobiology 3 © 2016 Macmillan Publishers Limited. All rights reserved
