recombinant DNA technology enzymes
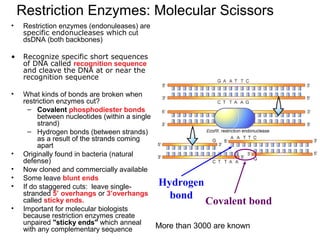
- 1. Restriction Enzymes: Molecular Scissors • Restriction enzymes (endonuleases) are specific endonucleases which cut dsDNA (both backbones) • Recognize specific short sequences of DNA called recognition sequence and cleave the DNA at or near the recognition sequence • What kinds of bonds are broken when restriction enzymes cut? – Covalent phosphodiester bonds between nucleotides (within a single strand) – Hydrogen bonds (between strands) as a result of the strands coming apart • Originally found in bacteria (natural defense) • Now cloned and commercially available • Some leave blunt ends • If do staggered cuts: leave single- stranded 5’ overhangs or 3’overhangs called sticky ends. • Important for molecular biologists because restriction enzymes create unpaired "sticky ends" which anneal with any complementary sequence Hydrogen bond Covalent bond More than 3000 are known
- 2. Restriction Enzymes called "restriction enzymes“ because restrict host range for certaincalled "restriction enzymes“ because restrict host range for certain bacteriophagebacteriophage bacterial" immune system": destroy any "non-self" DNAbacterial" immune system": destroy any "non-self" DNA methylase recognizes same sequence in host DNA and protects it by methylating it; restriction enzyme destroys unprotected = non- self DNA (restriction/modification systems) There are many examples of enzymes isolated from different sources which recognize the same target sequence. These are known as isoschizomers.Ex- MboI and Sau3AI
- 3. AGCCAG GATCCGGGCTGCAAGCGGTTAAG AATTCGTCGACGTCGACGAATTCTTAACCGCTTCCAGCCCGGATCCTGGCT GATCCGGGCTGCAAGCGGTTAAGAATTCTTAACCGCTTCCAGCCCG GATCCTGGCTAGCCAG AATTCGTCGACGTCGACG+ “sticky ends” AGCCAGAT ATCGGGCTGCAAGCGGTTAACAG CTGGTCGACGTCGACCAGCTGTTAACCGCTTCCAGCCCGATATCTGGCT ATCGGGCTGCAAGCGGTTAACAGCTGTTAACCGCTTCCAGCCCGAT AGCCAGATATCTGGCT + CTGGTCGACGTCGACCAG •“Sticky ends”: 5’ or 3’ over-hangs that allow the DNA to anneal even though it is not covalently bound •Help with the next step: ligation Most common restriction enzymes Blunt-end restriction enzymes No sticky ends Blunt vs Sticky Ends Digestion Digestion
- 4. Restriction endonuclease recognition sequences • Will occur randomly in any dsDNA • Statistically more often for e.g. 4-base cutters than 6-base cutters • If sites are known, we can use restriction enzyme cutting patterns to verify the identity of our DNA - called restriction mapping • Recognition sequences: usually 4 or 6 bases but there are some that are 5, 8, or longer • Recognition sequences are palindromes • Palindrome: sequence of DNA that is the same when one strand is read from left to right or the other strand is read from right to left– consists of adjacent inverted repeats • Recognition sequence may be interrupted or ambiguous – Acc I: GT(at/gc)AC – Bgl I: GCCNNNNNGGC – Afl III: ACPuPyGT
- 5. E.g. the recognition sequence for enzyme EcoRI (E. coli strain R1st enzyme found happens to be found) in this piece of DNA: 5’ -----G-A-A-T-T-C----- 3’ 3’ -----C-T-T-A-A-G----- 5’ Cutting the backbones between the red and blue nucleotides gives 2 nicks: 5’ -----G A-A-T-T-C----- 3’ 3’ -----C-T-T-A-A G----- 5’ The velcro bonds between the A’s and Ts break easily, giving 2 fragments with 5’ overhangs (sticky ends): 5’-----G 3’ + 5’ A-A-T-T-C----- 3’ 3’-----C-T-T-A-A 5’ 3’ G----- 5’
- 6. Note that whenever you have antiparallel, complementary sticky ends, they could potentially H-bond back together again: 5’-----G 3’ + 5’ A-A-T-T-C----- 3’ 3’-----C-T-T-A-A 5’ 3’ G----- 5’ back to: 5’ -----G A-A-T-T-C----- 3’ 3’ -----C-T-T-A-A G----- 5’ Only now there will be nicks between the red and blue bases (unless repaired by DNA ligase)
- 7. Restriction Endonucleases Type I- multisubunit, endonuclease and methylase activities, cleave at random nonspecific site > than 1,000 bp away Type II- cleave DNA within recognition sequence, require no ATP, most monomers Type III- Recognize specific 5-7 bp sequences multisubunit, endonuclease and methylase about 24-27 25 bp from recognition sequence TypeII b-
- 8. Origins of Restriction Enzymes • Naturally found in different types of bacteria • Bacteria use restriction enzymes to protect themselves from foreign DNA • Bacteria have mechanisms to protect themselves from the actions of their own restriction enzymes • Have been isolated and sold for use in lab work • Restriction enzymes are isolated from bacteria • Derive names from the bacteria • Genus- first letter capitalized • Species- second and third letters (small case) • Additional letters from “strains” • Roman numeral designates different enzymes from the same bacterial strain, in numerical order of discovery • Example: PstI Providencia stuartii 1st enzyme isolated EcoRI E Escherichia Co coli R R strain I first enzyme discovered from Escherichia coli R HindIII Haemophil us influenzae strain RD 3rd enzyme TaqI Thermus aqua 1st enzyme iso
- 9. Theoretical Basis Using Restriction Enzymes The activity of restriction enzymes is dependent upon precise environmental condtions: PH Temperature Salt Concentration Ions An Enzymatic Unit (u) is defined as the amount of enzyme required to digest 1 ug of DNA under optimal conditions: 3-5 u/ug of genomic DNA 1 u/ug of plasmid DNA Stocks typically at 10 u/ul
- 10. Conditions for activity Restriction Buffer provides optimal conditions • NaCI provides the correct ionic strength • Tris-HCI provides the proper pH • Mg2+ is an enzyme co-factor Why incubate at 37°C? • Body temperature is optimal for these and most other enzymes What happens if the temperature is too hot or cool? • Too hot = enzyme may be denatured (killed) • Too cool = enzyme activity lowered, requiring longer digestion time
- 11. •Endonucleases (or restriction enzymes) are enzymes which cut DNA at specific internal recognition sequences •Compare to exonucleases, which cut from one end •You must choose restriction sites that are available in the plasmid you are cloning into •They must not appear in your gene (silent mutation can remove unwanted sites in your designed gene) Choice of Restriction Sites/Enzymes Once you have your gene, you need to design a way to get it into your plasmid
- 12. •Restriction sites must exist only once in your plasmid •They must be in the correct position relative to the purification tag •Restrictions sites usually add extra residues to your gene product; make sure they are compatible with your peptide/protein •Some restriction sites are sub-optimal for cloning •Blunt end sites •dam and dcm methylation-affected enzymes Really Important Factors to Remember When Choosing Restriction Enzymes
- 13. dam Methylation N O N N O N N HO P P O O O O O O MeN O N N O N NH2 O P P O O O O O O Dam methylase •Dam methylase puts a methyl group on the nitrogen of 6th position of adenosine at the site: GATC •All of the E. Coli that we use generate DNA with dam methylation •Some enzymes only cut dam methylated DNA: eg DpnI •Some enzymes do not cut dam methylated DNA: eg XbaI http://www.neb.com/nebecomm/tech_reference/restriction_enzymes/dam_dcm_methylases_of_ecoli.asp
- 14. dcm Methylation http://www.neb.com/nebecomm/tech_reference/restriction_enzymes/dam_dcm_methylases_of_ecoli.asp Dcm methylase O P O O O O N N NH2 O O P O O O Me O P O O O O N N NH2 O O P O O O •Dcm methylase puts a methyl group on the carbon of 5th position of cytidine at the site: CCAGG and CCTGG •The enzyme we use most that can be affected by dcm methylation is SfiI •XL1-Blues and BL21s are both Dcm +
- 15. Application 1: Restriction Analysis • Using restriction enzymes to find out information about a piece of DNA • We can use restriction enzymes to find out – The size of a plasmid – If there are any restriction sites for a particular enzyme on a piece of DNA (ex. EcoRI) – How many restriction sites for a particular enzyme – Where the restriction sites are located
- 16. •Once you have your restriction enzymes chosen, it is time to design the final complete gene •The multiple cloning site (or whatever plasmid you are cloning into) should already have the 5’ portion of the gene intact (i.e. RBS, spacer, Met) • Sequences must be in frame NcoI BtgI 51 CTTTAATAAG GAGATATACC ATGGGCAGCA GCCATCACCA TCATCACCAC M G S S H H H H H H SacI AscI SbfI SalI NotI BamHI EcoRI EcoICRI BssHII PstI AccI HindIII 101AGCCAGGATC CGAATTCGAG CTCGGCGCGC CTGCAGGTCG ACAAGCTTGC S Q D P N S S S A R L Q V D K L A Application 2: Design of insert
- 17. Design of the Insert 71 ATGGGCAGCAGCCATCACCATCATCACCAC M G S S H H H H H H SacI AscI SbfI SalI BamHI EcoRI EcoICRI PstI AccI HindIII 101AGCCAGGATCCGAATTCGAGCTCGGCGCGCCTGCAGGTCGACAAGCTTGC S Q D P N S S S A R L Q V D K L A The gene we want: ggctgcgacagggcgagcccgtactgcggttaa G C D R A S P Y C G * BamHI PstI AGCCAGGATCCGAATTCGAGCTCGGCGCGCCTGCAGGTCGACAAGCTTGC S Q D P N S S S A R L Q V D K L A G C D R A S P Y C G * ggctgcgacagggcgagcccgtactgcggttaa AGCCAGGATCCGggctgcgacagggcgagcccgtactgcggttaaCTGCAGGTCGACAA Be aware of the amber stop codon: TAG Multiple cloning site
- 18. Design of the Insert Always check and re-check your sequence! ATGGGCAGCA GCCATCACCA TCATCACCAC AGCCAGGATCCGggctgcgacagggcgagcccgtactgcggttaaCTGCAGGTCGACAA atgggcagcagccatcaccatcatcaccacagccaggatccgggctgcgacagggcgagc M G S S H H H H H H S Q D P G C D R A S ccgtactgcggttaactgcaggtcgacaa P Y C G - L Q V D Everything looks good: in frame the whole way! Translate the whole gene
- 19. The wrong way to do it: AGCCAGGATCC ggctgcgacagggcgagcccgtactgcggttaaCTGCAGGTCGACAAGCTT atgggcagcagccatcaccatcatcaccacagccaggatccggctgcgacagggcgagcc M G S S H H H H H H S Q D P A A T G R A cgtactgcggttaactgcaggtcgacaagctt R T A V N C R S T S Frame shifted = garbage! Design of the Insert The gene is just inserted after the restriction site, which is out of frame with the plasmid-encoded start-codon/His- tag **Some plasmids, for whatever reason, have restriction sites out of frame with the translated gene**
- 20. Application 3: Restriction mapping • A restriction map is a map of known restriction sites within a sequence of DNA. Restriction mapping requires the use of restriction enzymes. In molecular biology, restriction maps are used as a reference to engineer plasmids or other relatively short pieces of DNA, and sometimes for longer genomic DNA. There are other ways of mapping features on DNA for longer length DNA molecules, such as mapping by Transduction (Bitner, Kuempel 1981).
- 21. Restriction mapping involves first cutting dsDNA with restriction enzymes (A, B, or both A and B) to produce predictably sized fragments: A B A A only: B only: A and B:
- 22. Constructing a restriction map for EcoRI and BamHI in a DNA fragment
- 24. Application: RFLP Analysis: •RF stands for Restriction Fragments. Those are the fragments that were cut by restriction enzymes. •L stands for Length, and refers to the length of the restriction fragment. •P stands for Polymorphisms, a Greek term for “many shapes”. The lengths of some of the restriction fragments differ greatly between individuals. RFLP = Restriction Fragment Length Polymorphism Molecular biologists have identified regions of the human genome where restriction fragment lengths are highly variable between individuals. Electrophoresis of these RFLP’s produce different patterns of DNA bands. With 3 billion base pairs in the human genome, however, RFLP analysis would produce a ‘smear’ of many similar sized fragments.