Lecture 4 winter 2012
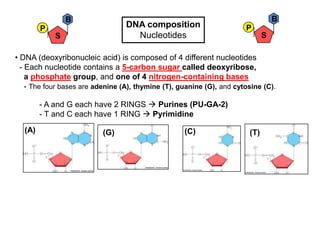
- 1. B B P DNA composition P S Nucleotides S • DNA (deoxyribonucleic acid) is composed of 4 different nucleotides - Each nucleotide contains a 5-carbon sugar called deoxyribose, a phosphate group, and one of 4 nitrogen-containing bases - The four bases are adenine (A), thymine (T), guanine (G), and cytosine (C). - A and G each have 2 RINGS Purines (PU-GA-2) - T and C each have 1 RING Pyrimidine (A) (G) (C) (T)
- 2. DNA composition Nucleotides • Closer look at deoxyribose sugar - Learn the numbering scheme - Base is connected to C1 of the sugar, Phosphate is on C5 - Notice C2 has an H and not an OH - C3 has a OH
- 5. DNA structure How are nucleotides connected to one another? • Nucleotides are connected via a phosphodiester bond - Between 5' phosphate group of 1 nucleotide and the 3’ OH group of a second nucleotide Each phosphate is connected to 2 sugars - One end of a polynucleotide will have a free 5’ phosphate (5’ end) and the other end will have an unbound 3’ OH group (3’ end)
- 6. DNA structure 3-D shape • Information known pre-1953 - DNA is a polynucleotide strand(s) that consists of nucleotides • Maurice Wilkins and Rosalind Franklin at King’s College (UK) (1952)
- 7. DNA structure 3-D shape • Watson, Crick, Franklin, and Wilkens model (1953) 1) DNA is a double-stranded polynucleotide that adopts a right-handed helix - The two strands are antiparallel (opposite directions) - 5' end of one interacts with 3' of the other 2) Bases lie flat in 3-D (perpendicular to axis)
- 8. DNA structure 3-D shape • Watson and Crick model of DNA 3. Outside of each strand (the backbone) - Consists of sugar and phosphate groups attached to one another as described previously 4. The bases face the inside and form the hydrogen bonds with the bases from the opposite strand. - Hydrogen bonds only form between A-T (2) and C-G (3) 5. Contains alternating major and minor grooves - Empty spaces in the 3-D structure of the double helix - Common location of protein binding A-T 2 H-bonds G-C 3 H-bonds
- 10. DNA structure Alternate forms of DNA • The type of DNA described by Watson and Crick and the type found in most normal cells is called B-DNA • However, DNA can adopt other 3-D shapes 1. A-DNA – Observed when water is removed from the DNA (dehydration/high-salt) - It is more compact than B DNA (11-12 bases/turn) - Still a right-handed helix - Bases are tilted upwards slightly (not flat) - Grooves are deeper, but narrower 2. Z-DNA – Observed when DNA is constructed to have mostly C-G base pairs - Has a spread-out, zig-zag shape - LEFT-HANDED helix - Major groove is not present
- 12. DNA structure Alternate forms of DNA • A and Z DNA have some biological relevance 1. A DNA and bacterial endospores - Some bacteria species (Gram positive) form protective endospores when environmental conditions become too harsh - Protected against heat, UV light, most harsh chemicals, drying, etc... 1. Remove most of their water and have a tough outer coat 2. Produce SASPs (spores associated proteins) that bind to the DNA and convert it from B to A - Tightens the DNA and protects it from damaging agents (e.g. UV light) 2. Z-DNA and the proteins that bind to them - Scientists have discovered cellular and viral proteins that have the ability to bind tightly to Z DNA (but not A or B DNA) - Cells wouldn’t have such proteins if Z DNA did not exist naturally - Now believe that many genes have stretches of sequence (GC rich) that adopt a Z like conformation (may be involved in transcription regulation)
- 13. Concept Check The antiparallel nature of DNA refers to: a. its charged phosphate groups. b. the pairing of bases on one strand with bases on the other strand. c. the formation of hydrogen bonds between bases from opposite strands. d. the opposite direction of the two strands of nucleotides.
- 14. Pathways of information transfer within the cell The Central Dogma
- 15. 10.4 Special Structure Can Form in DNA and RNA • Hairpin structure: in single strands of nucleotides, when sequences of nucleotides on the same strand are inverted complements, a hairpin structure will be formed. • When the complementary sequences are contiguous, the hairpin has a stem but no loop.
- 16. 10.4 Special Structure Can Form in DNA and RNA • RNA molecules may contain numerous hairpins, allowing them to fold up into complex structures.
- 17. RNA structure Basics • RNA/DNA similarities - Helical-shaped, polynucleotide chain that is composed of 4 types of nucleotides - Nucleotides are attached via phosphodiester bonds • RNA/DNA structural differences - RNA is mostly single-stranded (but can fold back on itself and form double-stranded regions) - RNA does not contain the base thymine. Instead, it contains a different pyrimidine called uracil (A-U hydrogen bond) - The sugar in the RNA nucleotides is RIBOSE (not deoxyribose) • RNA serves as the genetic material for a number of viruses - e.g. influenza, HIV, polio, yellow fever
- 18. Techniques for analyzing nucleic acids Absorption of UV light • Common techniques for measuring and analyzing DNA include: 1. Absorption of UV light (Spectroscopy) - DNA absorbs UV light with a wavelength of 260 nm very well - Other macromolecules (proteins, carbs, lipids) do not - Put sample into spectrophotometer and set to 260 nm - Measures amount of light absorbed/transmitted - More absorbance = more quantity of DNA - Can also be used to measure DNA purity - Proteins and other contaminants absorb light at 280 nm - Measure sample at 260 nm and 280 nm and get the ratio (Abs260/Abs280) - Example: Abs260 Abs280 Sample 1 0.75 1.5 Sample 2 1.5 1.5 Which would you want? Sample 3 0.5 0.1
- 19. Techniques for analyzing nucleic acids Centrifugation • Common techniques for measuring and analyzing DNA include: 2. Centrifugation - Centrifuge – Machine with an internal rotor that spins at very high speeds - The force generated from the spinning causes any particle with mass to move to the bottom of the tube - Can take a mixture of DNA, protein, carb, lipid and spin it in a centrifuge - Different components will separate out based on mass, shape, and density (ANALYSIS AND PURIFICATION) - Two major classes of centrifugation a) Sedimentation equilibrium centrifugation - A density gradient (usually CsCl) is created and mixture is applied - Mixture is spun for a fixed time - Molecules separate from one another based on their density - They move to their buoyant density and form a band in the tube
- 20. Techniques for analyzing nucleic acids Centrifugation • Common techniques for measuring and analyzing DNA include: 2. Centrifugation - Two major classes of centrifugation a) Sedimentation equilibrium centrifugation - Once done, pop hole in bottom of the tube and collect fractions - Measure each fraction for the presence of DNA or protein b) Sedimentation velocity centrifugation - This spins and measures how “fast” a molecule moves down a tube during centrifugation (analyzes and separates) - DNA/RNA shape and mass both influence how fast it moves - Expressed in Svedberg units (S) –used to calculate mass
- 21. Techniques for analyzing nucleic acids Denaturation and renaturation • Common techniques for measuring and analyzing DNA include: 3. Denaturation and renaturation - Can learn a lot about DNA by separating the two strands and allowing them to get back together - Heat DNA Hydrogen bonds between the two strands break and the 2 strands separate - No covalent (phosphodiester) bonds are broken - Called denaturation - Cool DNA Strands go right back together (forming proper hydrogen bonds) - Called renaturation or annealing a) Determining relative ratios of A/T and G/C - Remember: G and C form 3 hydrogen bonds A and T form 2 hydrogen bonds DNA with more G-C pairs will require hotter temps to denature
- 22. Techniques for analyzing nucleic acids Denaturation and renaturation • Common techniques for measuring and analyzing DNA include: 3. Denaturation and renaturation a) Determining relative ratios of A/T and G/C - Can plot the rate of separation with respect to temp. - Melting temperature (Tm) = Temp. at which half the DNA molecules have denatured Such analysis provides valuable info on the base composition of a piece of DNA (how many A, T, C, G) - A piece of DNA with a ton of G-C pairs will have a high melting temp., whereas a piece of DNA with fewer G-C, will have a lower melting temp. - Not about speed of denaturation - Its all about the temp!!!
- 23. Techniques for analyzing nucleic acids Denaturation and renaturation • Common techniques for measuring and analyzing DNA include: 3. Denaturation and renaturation b) Determining relative sequence similarity between two pieces of DNA - Take two different pieces of DNA and denature, mix, and allow to renature 5’ - DO THE DIFFERENT PIECES HYDROGEN BOND WITH EACH OTHER?? 3’ 5’ 3’ 3’ 5’ 3’ 5’ heat heat Allows to determine evolutionary relationships mix and cool - Does gene X from humans have a similar sequence as the same gene in apes? region of sequence similarity region of sequence difference
- 24. Techniques for analyzing nucleic acids Denaturation and renaturation • Common techniques for measuring and analyzing DNA include: 3. Denaturation and renaturation c) Determining sequence complexity - Can determine how complex a sequence is by denaturing DNA and looking to see HOW LONG it takes for the DNA to renature: - The more repetitive a sequence, the quicker it will renature ATATATATATATA CTGATGTCAAGT TATATATATATAT GACTACAGTTCA Which one would anneal faster if I heated them up? - This technique was critical in the identification of repetitive sequences in most of the human chromosomes (our chromosomes have lots of repeated sequences in between genes)
- 25. Techniques for analyzing nucleic acids Denaturation and renaturation • Common techniques for measuring and analyzing DNA include: 3. Denaturation and renaturation d) FISH (fluorescence in situ hybridization) - Design a single-stranded DNA probe that is specific for some DNA sequence (e.g. gene) - Tag the probe with a fluorescent dye - Add to cells (or chromosomes) of interest - Probe will anneal (renature) with its target sequence wherever it is located Location of fluorescence = Location of target - Major uses (for DNA): - Is the target gene present in the genome? - Find location of a gene on a chromosome - Is it where it is supposed to be? Abundance?
- 26. Techniques for analyzing nucleic acids Radioactivity • Common techniques for measuring and analyzing DNA include: 4. Radioactivity - Can tag nucleic acids with various radioisotopes - 32P is most common – also have radioactive C, N, or H - Tagging can be done in vivo or in vitro - Machines (e.g. scintillation counter) exist that can count how much radioactivity is present in a given sample - Can use to measure, follow, detect nucleic acids - Examples: 1) Hershey and Chase experiment 2) HSV-1 tracking - Tag a population of viral particles with radioactive genome - Infect an animal with that virus - Where is the radioactivity in that animal?
- 28. Techniques for analyzing nucleic acids Gel electrophoresis • Common techniques for measuring and analyzing DNA include: 5. Gel electrophoresis - Method of separating molecules in a mixture by adding the mixture to a semi- solid gel and applying an electric current through it - Allows for the separation of different DNA fragments based on their SIZE - Procedure a) The mixture of DNA fragments is added to a semi-solid gel (usually made of agarose) - Agarose – A large polysaccharide that is derived from seaweed b) Apply an electric current - Phosphates in DNA make it negatively charged. It will move towards the + electrode +
- 29. Techniques for analyzing nucleic acids Gel electrophoresis • Common techniques for measuring and analyzing DNA 5. Gel electrophoresis c) Agarose forms a big complex network - DNA fragments have to wiggle their way through it - Smaller the fragment, easier time of getting through the agarose, faster it will move SMALLER DNA FRAGMENTS WILL MIGRATE FASTER THAN LARGE FRAGMENTS - Visualization can also give indication of quantity. + small fragment
- 30. Benjamin A. Pierce GENETICS A Conceptual Approach FOURTH EDITION CHAPTER 11 Chromosome Structure and Transposable Elements © 2012 W. H. Freeman and Company
- 31. Chapter 11 Outline 11.1 Large Amounts of DNA Are Packed into a Cell, 292 11.2 Eukaryotic Chromosomes Possess Centromeres and Telomeres, 299 11.3 Eukaryotic DNA Contains Several Classes of Sequence Variation, 301 11.4 Transposable Elements Are DNA Sequences Capable of Moving, 303 11.5 Different Types of Transposable Elements Have Characteristic Structures, 308 11.6 Transposable Elements Have Played an Important Role in Genome Evolution, 314
- 32. 11.1 Large Amounts of DNA Are Packed into a Cell • Supercoiling – Positive supercoiling Fig. 11.2 (b) – Negative supercoiling Fig. 11.2 (c) – Topoisomerase: The enzyme responsible for adding and removing turns in the coil.
- 33. Relaxed DNA: Every 100 BPs Have 10 turns. LESS turns than you Expect. Ex: 100 BPs Having 5 turns More turns than you Expect. Ex: 100 BPs Having 20 turns
- 35. 11.1 Large Amounts of DNA Are Packed into a Cell • Chromatin Structure – Euchromatin: what gets transcribed – Heterochromatin: never gets transcribed! – Histone proteins: (+) Charged proteins that stick to (-) charged DNA and form nucleosomes
- 36. Concept Check 2 Neutralizing their positive charges would have which effect on the histone proteins? a. They would bind the DNA tighter. b. They would separate from the DNA. c. They would cause supercoiling of the DNA.
- 37. 11.1 Large Amounts of DNA Are Packed into a Cell • Chromatin Structure • The nucleosome • Chromatosome • Linker DNA • High – order chromatin structure – Fig. 11.4, 11.5
- 38. 11.1 Large Amounts of DNA Are Packed into a Cell
- 41. 11.1 Large Amounts of DNA Are Packed into a Cell • Chromatin Structure – Changes in chromatin structure • Chromosomal puffs – Fig. 11.7 • Telomere sequences – Fig. 11. 12
- 42. Place of high transcription
- 44. 11.4 Transposable Elements Are DNA Sequences Capable of Moving • General characteristics of transposable elements:
- 45. Benjamin A. Pierce GENETICS A Conceptual Approach FOURTH EDITION CHAPTER 12 DNA Replication and Recombination © 2012 W. H. Freeman and Company
- 46. Chapter 12 Outline • 12.1 Genetic Information Must be Accurately Copied Every Time a Cell Divides, 322 • 12.2 All DNA Replication Takes Place in a Semi-conservative Manner, 322 • 12.3 Bacterial Replication Requires a Large Number of Enzymes and Proteins, 330 • 12.4 Eukaryotic DNA Replication Is Similar to Bacterial Replication but Differs in Several Aspects • 12.5 Recombination Takes Place Through the Breakage, Alignment, and Repair of DNA Strands.
- 47. one chromosome (unduplicated) DNA replication When and why? one chromosome (duplicated) • Review of Bio I - Cell division requires a previous DNA replication (if not, see picture below) - Eukaryotic cells do this in S phase of interphase - Bacterial cells do so before binary fission No rep 46 23 23 11 12 11 12 further reduction + rep 46 46 46 46 46 46 46 NO reduction • Our 46 chromosomes are collectively composed on 6 billion base pairs - Practical example: Typing out in font 10 Would fill 1200 textbooks - Our cells can copy all 46 chromosomes in Remember the ~6 hours. Lots of errors?? whispering game - NO!! The error rate is only What happens to the about 1 per 10 million nucleotides info after being copied a few times?
- 48. 12.2 All DNA Replication Takes Place in a Semiconservative Manner • Up until Watson and Crick made their discovery, scientists had no idea how DNA was replicated during S phase - Their model provided a starting point for better understanding this process • Some possible mechanisms of DNA replication: (1) Semiconservative – Each strand serves as a template for a new strand - Each new double-stranded DNA molecule contains 1 old strand and 1 new strand (2) Conservative – Would result in 1 completely old and 1 new double-stranded DNA molecule. (3) Dispersive – Would produce two DNA molecules with sections of both old and new DNA interspersed along each strand
- 49. 12.2 All DNA Replication Takes Place in a Semiconservative Manner • Meselson and Stahl’s Experiment: – Two isotopes of nitrogen: • 14N common form; 15N rare heavy form • E.coli were grown in a 15N media first, then transferred to 14N media • Cultured E.coli were subjected to equilibrium density gradient centrifugation: – Fig. 12.3
- 50. How is DNA replicated? Meselson-Stahl experiment -1958 • They grew E.coli first in medium containing "heavy" nitrogen (15N) - 15N is a stable isotope of nitrogen that has 1 more neutron than the normal 14N - The extra neutron makes it a little more dense - All DNA bases initially contain the heavy nitrogen (both strands will be heavy) • They then switched the bacteria to medium containing lighter 14N and kept them there for a few cell divisions (generations) • They took bacteria at each time point, extracted the DNA, and separated the DNA on a density gradient (sedimentation equilibrium centrifugation) - DNA strands containing 15N are more dense than those containing 14N - They will move to a different part of the tube
- 51. How is DNA replicated? Meselson-Stahl experiment -1958 • After the first generation in 14N, would be expected? switch to Grown in 14N 15N If 15N/14N semiconservative 15N/15N or dispersive OR semi disper switch to Grown in 14N 15N If 14N/14N conservative 15N/15N 15N/15N
- 52. DNA replication is semiconservative! Meselson-Stahl experiment -1958 Animation http://highered.mcgraw-hill.com/sites/0072437316/student_view0/chapter14/animations.html# • After a second generation in 14N, what would be expected? 14N If 15N/14N 15N/14N 14N) (more dispersive 14N 14N/14N If 15N/14N 15N/14N semiconservative
- 54. DNA replication Overview • Overview of semiconservative DNA replication - Two parental strands are unwound and separated - Each of those serves as a template for a new, complimentary strand - End result = Two DNA molecules, each containing 1 parental and 1 new strand • We will first discuss how DNA replication proceeds in bacteria and then discuss how it occurs in more complex eukaryotic cells - Generally the same process in both cell types (as described above)
- 55. 12.2 All DNA Replication Takes Place in a Semiconservative Manner • Modes of Replication – Replicons: Units of replication. • Replication origin – Theta replication : circular DNA, E.coli; single origin of replication forming a replication fork, and it is usually a bidirectional replication. – Rolling-circle replication: virus, F factor of E.coli; single origin of replication.
- 56. DNA replication in bacterial cells Step 1: Initiation • Structure of the bacterial chromosome - Single, circular chromosome - Adopts a supercoiled state (which makes it more compact) • Bacterial DNA replication (also called Theta replication) relaxed supercoiled (1) Initiation - Does replication start anywhere on the chromosome or at specific sites? ANSWER: At specific sites called origins of replication - Bacteria have only a SINGLE origin of replication (DNA replication starts at one place in the chromosome - The origin of replication in the E.coli chromosome is called oriC - OriC (and other prokaryotic origins of rep.) contain binding sites for an initiator protein called DnaA - A complex of DnaA proteins bind to the chromosome at oriC and causes a slight “bend” to be introduced
- 57. DNA replication in bacterial cells Step 2: Unwinding the 2 strands • Bacterial DNA replication (also called Theta replication) (2) Unwinding a) Bending of the DNA at this point creates tension and causes a small replication “bubble” to form - This is a small region of strand separation b) The bubble serves as a binding site for 2 other proteins called DnaB and DnaC - These proteins use ATP to further break hydrogen bonds between the strands and separate them - DnaB and C are types of helicases - Helicase = Type of enzyme that uses ATP in order to denature DNA c) Proteins called single-strand-binding proteins (SSB proteins) bind to the individual strands and keep them apart
- 58. DNA replication in bacterial cells Step 2: Unwinding the 2 strands • Bacterial DNA replication (also called Theta replication) theta (2) Unwinding d) Unwinding the strands at oriC produces tighter supercoiling ahead of the replication fork - This tension needs to be relieved or the strands may break - An enzyme called DNA gyrase (a type of topoisomerase) binds to the chromosome “ahead” of the replication fork and relieves the tension - They introduce small cuts in the strands, unwind, and reseal Notes on unwinding: - Proceeds in both directions (aka bidirectional) - Produces a theta looking structure (see pict above) - It is unwound a little at a time - Unwound a little, new strands made, unwound a little more, .....
- 59. DNA replication in bacterial cells Step 3: Primer synthesis • Bacterial DNA replication (also called Theta replication) (3) Primer synthesis - The enzyme that makes a new DNA strand X from the template (called DNA polymerase III) can’t start the new strand from scratch - It only can add nucleotides to an existing 3’ OH group (It needs help getting things started) - What does the cell do? 1. Once the strands are separated, an enzyme called primase creates a short (5-10 nucleotides) RNA chain called a primer - This primer is complimentary to the template 2. DNA polymerase III can then come in and create a new strand by extending from the primer (called elongation) NO PRIMERS, NO DNA REPLICATION!!!
- 60. DNA replication in bacterial cells Step 4: Synthesis of new DNA strands • Bacterial DNA replication (also called Theta replication) (4) Elongation (synthesis of new strands) - First, let’s talk more about DNA polymerase III - Arthur Kornberg discovered DNA polymerases in 1956 (actually discovered a different DNA pol – discussed later) - Arthur’s son Thomas Kornberg discovered DNA pol III in 1970 - Properties of DNA polymerase III a) Only adds new nucleotides onto the 3’ end of a growing strand - Makes a new DNA strand in the 5’ 3’ direction - A dNTP is brought in, pyrophosphate (P-P) is removed - It reads what is on the template strand and adds in complementary nucleotide - Reads T, adds A; reads G, adds C b) Again, it can’t start a strand from scratch. It only can extend an existing strand
- 61. DNA replication in bacterial cells Step 4: Synthesis of new DNA strands • Bacterial DNA replication (also called Theta replication) (4) Elongation (synthesis of new strands) - Properties of DNA polymerase III c) It has 3’5’ exonuclease activity - Has the ability to proofread itself and correct its mistakes - If it detects that it accidentally inserted the wrong nucleotide, it will back up, cut it out, and put the correct one in - Analogy: Backspace key on the keyboard d) Structure - Composed of many different functional subunits 1. Three subunits make up the core enzyme α synthesizer – polymerization of nucleotides ε proofreader – corrects mistakes made θ stimulator – stimulates ε to do its job 2. β clamp – Sliding clamp structure that locks the whole complex onto the DNA strands - Prevents the above core enzyme from falling off
- 62. DNA replication in bacterial cells Step 4: Synthesis of new DNA strands • Bacterial DNA replication (also called Theta replication) (4) Elongation (synthesis of new strands) - Properties of DNA polymerase III d) Structure - Composed of many different functional subunits 3. Tau (η) subunit - Tethers two DNA pol. III together (creates a dimer) 4. Gamma (γ) loader complex – Helps to load the whole enzyme onto the DNA template at the replication fork (lagging strand) - How does DNA pol. III actually make new strands of DNA? - Remember the 2 strands of DNA are arranged in opposite (antiparallel) orientations 5’ 3’ 3’ 5’ As new strands are produced, they must also follow this antiparallel rule (Two interacting DNA strands must always run in opposite directions)
- 63. DNA replication in bacterial cells Step 4: Synthesis of new DNA strands • Bacterial DNA replication (also called Theta replication) (4) Elongation (DNA synthesis) - Now, for the actual steps 5’ 3’ - Remember: Starts with localized DNA unwinding at 5’ 3’ oriC and the addition of RNA primers (in 3’ green) by primase 5’ - Notice where they are 5’ 3’ placed on the 2 strands 5’ 3’ 3’ - DNA pol III then adds 5’ nucleotides onto the 3’ end of each primer (in red) 3’ - Synthesis continues in the 5’ 5’ 3’ 5’ 3’ direction 5’ - Notice one is moving 3’ 3’ towards the replication 5’ fork and the other away Note: Only 1 replication fork is shown
- 64. DNA replication in bacterial cells Step 4: Synthesis of new DNA strands • Bacterial DNA replication (also called Theta replication) (4) Elongation (DNA syn.) - Now, for the actual steps - One strand is being made continuously as DNA pol III moves towards the replication fork (this is called the LEADING STRAND) - The other strand is made discontinuously (moving away from the replication fork) and is initially composed of small 3’ 5’ fragments called 5’ 3’ Okazaki fragments 5’ (this is called the 3’ 3’ 5’ 3’ LAGGING STRAND) 5’ Prediction of next steps: 5’ 3’ 1. Can’t have pieces of 3’ 5’ RNA in our chromosome + 2. One of the new strands 5’ 3’5’ 3’ 5’ 3’5’ 3’ is in pieces 3’ 5’
- 65. DNA replication in bacterial cells Step 5: Primer removal and ligation http://highered.mcgraw-hill.com/sites/0072437316/student_view0/chapter14/animations.html# • Bacterial DNA replication (also called Theta replication) (5) Primer removal and ligation - Another type of DNA polymerase (DNA polymerase I) comes in and removes the RNA primers from both the leading and lagging strands (it has 5’3’ exonuclease activity) and fills in the gaps with DNA (it also has 5’3’ polymerization ability) - It uses the end of 5’ 3’ each Okazaki fragment 3’ 5’ as a starting point + 5’ 3’ 5’ 3’ 5’ 3’ 5’ 3’ - Does both in 1 step! DNA pol I 3’ 5’ -The enzyme DNA ligase then seals the spaces 5’ 3’ between each Okazaki 3’ 5’ fragment on the lagging 3’ strand 5’ 3’ 5’ - Creates the last phosphodiester bond 5’ 3’ DNA ligase 3’ 5’
- 66. DNA replication (GENERAL) Proofreading and error correction • The proofreading ability (3’5’ exonuclease activity) of the DNA polymerases prevents massive amounts of errors from being introduced into the genome - How do we know this? - Removal of the 3’5’ exonuclease activity from E.coli DNA pol III results in a large increase in the error rate of the enzyme - MUTATIONS ARE INTRODUCED into newly made DNA strands • Why is preventing these mutations important? - Two words: DEATH and CANCER!! - Mutation of DNA often leads to cell death because if the wrong gene is crippled, the cell can’t function properly - Worse yet, mutation of specific genes can abnormally lead a cell to divide uncontrollably and prevent the cell from dying properly TUMOR PREVENTING MUTATIONS IS CRITICAL!!!
- 67. DNA replication in eukaryotic cells Similarities and differences to bacteria • DNA replication in eukaryotic cells is very similar to what occurs in bacterial cells - DNA unwinding at origin of replication, 2 replication forks are formed, RNA primers are made, and DNA polymerases make new strands on leading and lagging strands • However, differences between bacterial and eukaryotic genomes leads to some differences in how the DNA is replicated - Bacteria: Single, circular chromosome (Avg size: 4.6 x 106 base pairs) Humans: Multiple, linear chromosomes (Total size: 3.3 x 109 base pairs) • Difference 1: Eukaryotic chromosomes have multiple origins of replication - Example: Copying your textbook - Option A - You photocopy it by yourself from start to finish - Option B – You and 30 of your friends each copy a chapter Eukaryotic genomes are gigantic. Need to start replication at multiple sites in order to complete it in a timely manner
- 68. DNA replication in eukaryotic cells Similarities and differences to bacteria • Difference 1: Eukaryotic chromosomes have multiple origins of replication - How do our cells recognize all of these origins of replication and how do they keep from accidentally copying the same thing twice (before mitosis)? 1) In order to function, origins must be “licensed” or approved for replication by origin recognition complexes (ORCs) - ORCs function to recruit helicase, SSB protein, DNA pols, etc. - No ORC binding, no replication - After replication, ORCs come off 2) Unreplicated DNA is coated with MCM proteins (helicases) - Its one way that the cell has of distinguishing between replicated and unreplicated DNA - No MCM proteins, no replication - Following replication, MCM proteins come off the DNA and are prevented from going back on
- 69. DNA replication in eukaryotic cells Similarities and differences to bacteria • Difference 2: Eukaryotic DNA polymerases - Bacteria have at least 5 different DNA polymerases - Already discussed DNA pol I and III. DNA pol II, IV, and V are thought to function in DNA repair - At least 13 different DNA polymerases exist in eukaryotic cells - Some of the “important” ones are as follows: 1/2. Polymerase α (Pol α) and Pol δ (delta) – Major polymerases involved in nuclear DNA replication - Pol α – Two of its 4 subunits function as the primase - Adds primer to leading and lagging strand templates and starts extending them - This enzyme has low processivity and falls off of the templates after a short time - Pol δ – Replaces pol α and finishes the job (lagging strand) - Has high processivity and 3’5’ exonuclease activity (pol α does not) This is called polymerase switching 3. Pol ε – Similar abilities as pol δ, may be major leading strand polymerase (Still debated!!)
- 70. DNA replication in eukaryotic cells Similarities and differences to bacteria • Difference 2: Eukaryotic DNA polymerases - At least 6 different DNA polymerases exist in eukaryotic cells 4. Pol γ – Functions to replicate mitochondrial DNA - Mitochondria contain their own 16, 500 base pair, circular genome - Derived from bacteria?? - This DNA contains genes that are required for life - Most function in oxidative phosphorylation - Passed from mom to child (sperm don’t have lots of mitochondria) - Note: Chloroplasts also contain their own DNA and utilize a different DNA pol for DNA replication (still not fully characterized) 5-13. Many other eukaryotic DNA polymerases play specialized roles in DNA repair - You don’t need to know them
- 71. DNA replication in eukaryotic cells Similarities and differences to bacteria • Difference 3: Removal/assembly of histones - Eukaryotic nuclear DNA is tightly coiled around proteins called HISTONES (unlike bacterial DNA) - Remember: Chromosomes are DNA + protein - Histones are very basic proteins (+ charged) that interact with the negatively-charged DNA - Need to alter histones in order to make the DNA accessible for replication - Histones are typically altered by adding an acetyl group (called acetylation) - Adding an acetyl group (COCH3) neutralizes their positive charge, causes them to lose their attraction for DNA - DNA loosens up and is able to be replicated - After replication, histones are deacetylated and go back onto the DNA (and new histones are made)
- 72. DNA replication in eukaryotic cells Similarities and differences to bacteria • Difference 4: Chromosome ends/telomeres - Remember: Bacterial chromosomes are circular and eukaryotic chromosomes are linear - Remember: DNA polymerases can only add on to an existing 3’ end (all need a head start) - Remember: What does DNA pol I do in bacteria? - Removes the RNA primers and fills in the gaps with DNA (using the ends of the Okazaki fragments as a starting point) 5’ 3’ - Problem: Once DNA pol I removes 3’ 5’ the primer at the absolute end of end of the chromosome on the lagging chromsome strand, there is no free 3’ end 5’ 3’ ahead of it 3’ 5’ - In theory, our chromosomes 5’ 3’ should shorten each time they 3’ 5’ are replicated - Would lose important genes 5’ 3’ very quickly DEATH!! 3’ 5’
- 73. DNA replication in eukaryotic cells Similarities and differences to bacteria • Difference 4: Chromosome ends/telomeres - Another view of this problem - Solution to the problem: The end of our chromosomes (called telomeres) are modified to prevent the loss of important genes - An enzyme called telomerase is activated and adds >1,000 copies of a repeated sequence (e.g. TTAGGG) onto the ends of each chromosome - It is only active in gametes, embryos, and stem cells TTAGGGTTAGGGTTAGGGTTAGGGTTAGGG - Telomerase has a RNA template in a pocket that binds to the chromosome end - Uses this to keep adding more copies of the repeated sequence onto the telomere - Again, it is normally only active in sperm/eggs and in early embryos
- 74. DNA replication in eukaryotic cells Similarities and differences to bacteria • Difference 4: Chromosome ends/telomeres - Telomeres still shorten each time the DNA TTAGGGTTAGGGTTAGGGTTAGGGTTAGGG is replicated (problem wasn’t truly solved) - Instead of losing important genes, TTAGGGTTAGGGTTAGGGTTAGGG chromosomes only lose a portion of the TAGGGTTAGGGTTAGGG “garbage” repeated sequence - These repeated sequences at our telomeres also function to protect chromosomes by: 1) Serving as binding sites for protective proteins - Some enzymes love to degrade DNA by starting at ends (free ends are bad) - Repetitive sequences help recruit proteins to the telomeres - These proteins bind to the telomere and block the “bad” enzymes from destroying the chromosomes 2) Prevent chromosomes from fusing - If you cut chromosomes into small pieces and mix them, they tend to randomly fuse together - Repeated sequences prevent chromosomes from randomly fusing together
- 75. DNA replication in eukaryotic cells Similarities and differences to bacteria • Difference 4: Chromosome ends/telomeres - The more a cell divides, the shorter the telomeres get (remember: telomerase is not normally active in somatic cells) - As cells age, the telomeres get shorter and shorter - Eventually the repeated sequence runs out and important genes in the chromosomes get removed TTAGGGTTAGGGTTAGGGTTAGGGTTAGGG - Once this happens, the cell dies! TTAGGGTTAGGGTTAGGGTTAGGG TAGGGTTAGGGTTAGGG - Telomere shortening acts as a cell clock GGGTTAGGG - Does telomere shortening lead to human aging? - Possibly. Elderly people have shorter telomeres as do individuals with some aging diseases - Not all tissues are “aged” by shortened telomeres cell death
- 76. Replication - Recapitulating…. • DNA Polymerases Problems: Activities that are not inherited in the DNA polymerase itself. - There are certain polymerases that are specialized for different functions. - Not all DNA polymerases can replicate the genome - DNA Pol I in bacteria is a repair pol: Has 3 activities: 1) Polymerase activity 2) Proofreading exonuclease that goes 3’5’ 3) Exonuclease that chews-up the strand ahead in the 5’3’ direction - The structures of polymerases are conserved (they are ancient!) - Polymerases can’t start the polymer without a primer (needs a free 3’OH) - they can only synthesize in ONE direction! that creates a problem that is solved by a molecular machine called the replication fork (which includes ~10 different proteins that need to cooperate to replicate DNA) How does replication happen: • 1st problem: How to get the templates; how to separate the DNA strands? - Helicases unwind the double-stranded DNA. - Helicase loading triggers the assembly of the replication fork machine. dnaB helicase uses ATP hydrolysis to pry the strands apart (~1ATP/3 nucleotides). • 2nd problem: ssDNA is a signal for induction of cell death (EUK), and is unstable. • There are ss-nucleases floating around that can chew-it-up! Also, hairpins can block synthesis. so how do you mask the ssDNA? need stabilizing proteins (SSBP in Prok; or replication prot.A in Euk) - ss stabilizing proteins do not use ATP to bind. - creates a filament that is a better substrate for DNA polymerase (better than naked DNA). (Probably allosteric regulation on the polymerase)
- 77. Replication - Recapitulating…. • DNA Polymerase Problems: Activities that are not inherited in the DNA polymerase itself. • 3nd problem: Polymerase can’t start synthesis without a 3’OH. It needs a primer! First, helicase opens up the DNA, RNA primase is sitting on the fork (attracted by SSB) and takes care of the primer job. It lays a primer (10 nucleotides long) at the beginning of the leading strand and it lays primers every ~1000 nucleotides (in prokaryotes) on the lagging strand. There is no proofreading in primase (no backwards motion), so those primers have lots of errors. Then primase hands the primer to the polymerase (via clamp affinity). file:///E:/media/ch12/Animations/1201_replication.html file:///E:/media/ch12/Animations/1202_bidirectional_replic.html • 4th problem: DNA gets supercoiled (entangled) as it unwinds • during replication. Enzymes relieve the supercoiling: gyrases in prokaryotes; topoisomerases (type I and type II) in eukaryotes. they control the topology (# of turns) of the chromosomes. So… PROK: Here is the structure: 2 copies of DNA pol III bound together by the tau unit (holoenzyme). DNA Polymerase III: has (a) the polymerase subunit, (b) the proofreading subunit, (c) the exonuclease subunit, EUK: (a) DNA Pol a has the primase activity (30 – 40 nucleotides); (b) DNA Pol d polymerises the lagging strand; (c) DNA Pol e polymerises the leading strand. the changing of a to d and e is called polymerase switching.
- 78. Replication - Recapitulating…. • DNA Polymerases Problems: Activities that are not inherited in the DNA polymerase itself. • 4th problem: polymerase actually has low affinity to the ssDNA! solution: Use a beta clamp to keep the polymerase on the ssDNA. Clamp loader (does the ring-and-string magic): gamma complex in prokaryotes and replication factor C (made-up of A, B, C, D, E) in eukaryotes. The clamp loader uses ATP to load the clamp on the DNA. http://www.youtube.com/watch?v=-ie4dxx10Ww&feature=related (heavy accent, but good explanation…) The clamp itself is shaped like a donut and can not “magically” get into the ssDNA. clamp: beta in prokaryotes. The prokaryotic beta clamp (shown in the pic) recognizes the gamma complex clamp loader, then grabs the ssDNA strand. Qt: What does the clamp loader recognize? clamp: PCNA in eukaryotes. The eukaryotic DNA sliding clamp that keeps DNA polymerase engaged at a replication fork is called proliferating cell nuclear antigen (PCNA), is loaded onto the 3′ ends of primer DNA through its interaction with a heteropentameric protein complex called replication factor C (RFC) – the clamp loader. http://www.youtube.com/watch?v=HN7FQwogusY&feature=related So…. In prokaryotes: The subunits that hold the complex on each DNA strand are the beta/sliding clamps: there are lots of +charges on the clamps, so DNA levitates in the donut. Once the clamp goes in by association w/ the RNA primer, it does not come off the leading strand. On the lagging strand, the clamp associates with the RNA primer just as it did on the other strand, then attracts Polymerase III to synthesize the DNA. When beta senses the adjacent Okasaki fragment of DNA, it falls off, therefore Pol III looses its grip and falls off too. Pol III then finds the next beta clamp on the next RNA primer and joins for the synthesis again.
- 79. Replication - Recapitulating…. • DNA Polymerases Problems: Activities that are not inherited in the DNA polymerase itself. • 5th problem: how to remove the RNA primers? solution: in prok: DNA Pol I binds and replaces the RNA for DNA. Pol I has proofreading capacity. Now… At the nick there is a 3’OH and a 5’ phosphate. We could have an electrophilic attack from the OH onto the –P, but there is no “leaving group,” so the electrons would just go back and forth. We can’t seal the nick if the –P is not activated. DNA ligase first activates the –P (using ATP), then catalises the attack of the 3’OH on the activated –P. Now the nick is sealed! In euk: RNaseH1 endonuclease, cleaves RNA primer. Proofreading (with help of other subunits). DNA ligase I ligates Okazaki fragments, binds to PCNA. (Note: Unlike E. coli, which has only one DNA ligase, there are at least four known human DNA ligases). Uses ATP as an energy cofactor.
- 80. Replication - Recapitulating…. euk Two DNA polymerases are involved in eukaryotic replication DNA polymerase d has no primase activity and is thought to be the polymerase that synthesizes the leading strand. DNA polymerase a has associated primase activity and is thought to be the polymerase that synthesizes the lagging strand. DNA Synthesis at the Origin Additional factors: PCNA (proliferating cell nuclear antigen) DNA helicase Replication factor C OTHERS Replication of Nucleosomes
- 82. Transcription - Recapitulation • All genes (eukaryotic and prokaryotic) have a unique region (promoters) of DNA sequence upstream of (before) the transcriptional start site that serves as a binding site for the RNA polymerase enzyme • General functions of promoters: 1. Provides specificity – Each gene has a unique promoter sequence 2. Tell RNA polymerase where to start (they are usually at the beginning) - No promoter (or mutated promoter), no transcription 3. Indicate which strand will be transcribed and the direction of transcription • Most promoters share a few common sequences called consensus sequences Prokaryotic promoters share 2 main types of consensus sequences - TATA box (aka Pribnow box) (-10-15), sequence is usually TATAAT - TTGACA – found 35 nucleotides upstream • RNA polymerase enzyme makes direct contact with the promoter at these consensus sequences • Unique seq. within and around promoters help determine rates of transcription Prokaryotic cells only produce 1 type of RNA polymerase to produce all of the mRNA, tRNA, and rRNA in the cell. • Prokaryotic RNA polymerase structure: - 2 alpha (α), 1 beta (β), 1 beta prime (β'), and 1 sigma (ζ) subunit - Together they have the basic catalytic function of producing new RNA - ζ factor controls binding to the promoter (PROMOTER RECOGNITION) - After initial binding has been successful, the ζ factor comes off the core - Bacteria make many different types of sigma factors - Different sigma factors recognize different promoter sequences.
- 83. Transcription - Recapitulation • Prokaryotic RNA polymerase structure: - β, β’, α subunits interact and form the CORE ENZYME - Together they have the basic catalytic function of producing new RNA - ζ factor controls binding to the promoter (PROMOTER RECOGNITION) - After initial binding has been successful, the ζ factor comes off the core - Bacteria make many different types of sigma factors - Different sigma factors recognize different promoter sequences • Once it binds to the specific promoter, RNA polymerase positions itself over the transcriptional start site based on its distance from the consensus sequences - No specific “start signal” exists • RNA pol unwinds the DNA near the start site (transcription bubble created). • RNA pol continues producing a complimentary RNA molecule until it transcribes a terminator signal. Two major prokaryotic transcriptional terminator signals exist - Both must: (a) Slow down the RNA polymerase; (b) Weaken the interaction between the DNA and RNA in the bubble. Properties of the two major prokaryotic terminator signals 1. Rho-independent terminators - The DNA sequence contains an inverted repeat followed by a string of 6 adenines. Following their transcription, the inverted repeats form Hydrogen bonds with each other within the RNA and form a hairpin loop - Loop formation causes the RNA pol to pause - The pause combined with weak hydrogen bonding between 6 straight A-U pairs cause the RNA to totally fall off of the DNA template
- 84. Transcription - Recapitulation • (… two major prokaryotic terminator signals) 2. Rho-dependent terminators - Also contain inverted repeats that cause hairpin loop formation in the new RNA. Slows down the polymerase - RNA contains a binding site for the Rho protein. Rho moves towards the 3’ end - Acts as a RNA/DNA helicase. Rho is ATPase and helicase. - Many prokaryotic genes are organized in operons (genes in tandem, in similar pathway). Eukaryotic transcription follows the same basic steps of prokaryotic transcription - Activation, initiation, elongation, termination - Major differences exist in how they accomplish the above steps • General features of transcriptional activation is similar in prokaryotic and eukaryotic cells - Some genes are constituitivelly expressed, others are regulated - The exact nature of the signals that activate transcription can be very different • Initiation – Making the DNA accessible. Eukaryotic DNA is tightly coiled around proteins called histones (and some nonhistone proteins) - DNA is negatively charged (phosphate groups), histones are very positively charged - A histone octamer (8 histone proteins in a complex). Each group is called a nucleosome • DNA is freed from histones in 2 major ways: 1) Histone acetylation (HATs) add an acetyl group (CH3CO) to histones. This neutralizes their positive charge and causes them to lose their attraction for DNA. - Occurs only in the area to be transcribed (it is specfic!) Deacetylases remove acetyl groups after transcription 2) Chromatin remodeling - Some proteins move nucleosomes around without directly modifying histones.
